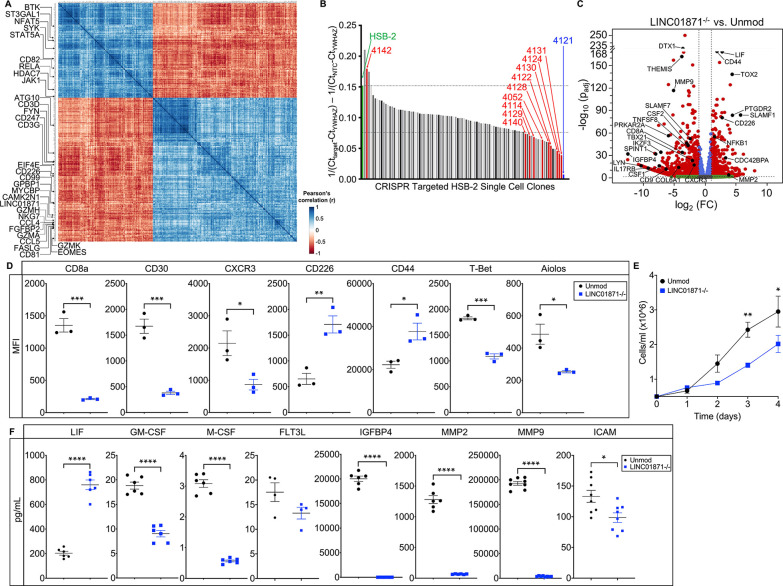
Figure 3.
Loss of LINC01871 disrupts basal expression of genes involved in immune cell regulation. (A) LINC01871 correlation analysis of normalised RNA-seq data from the SjDRo−-only primary expression matrix (r≥0.7 or ≤−0.6; p<0.05). Transcripts implicated in immune function or SjD pathology are indicated. (B) Quantitative PCR screen of LINC01871 expression in HSB-2 single-cell clones after CRISPR-targeted deletion of LINC01871. (C) Differentially expressed transcripts from the RNA-seq analysis of HSB-2 clone 4121 (hereafter LINC01871−/−) relative to HSB-2 parental cell line. Y-axis shows the −log10 of the FDR-adjusted p value (padj); x-axis shows the log2 of the fold change (FC). Black dots indicate independently replicated transcripts of interest. (D) Levels of indicated surface or intracellular proteins, reported as mean fluorescence intensity (MFI), in HSB-2 parental cells (black) and LINC01871−/− cells (blue); n=3; unpaired t-test where *p<0.05, **p<0.01 or ***p<0.001. (E) Growth curve analysis of HSB-2 parental cells (black) and LINC01871−/− cells (blue) from 0 to 96 hours; n=3; unpaired t-test where *p<0.05 or **p<0.01. (F) Concentration of indicated secreted protein in supernatant collected from HSB-2 parental cells (black) and LINC01871−/− cells (blue) at 96 hours; n>6; unpaired t-test where *p<0.05 or ****p<0.0001. FDR, false discovery rate; RNA-seq, RNA sequencing; SjD, Sjögren’s disease; SjDRo−, anti-Ro negative SjD.