Abstract

Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the virus that causes COVID-19, continues to evolve resistance to vaccines and existing antiviral therapies at an alarming rate, increasing the need for new direct-acting antiviral drugs. Despite significant advances in our fundamental understanding of the kinetics and mechanism of viral RNA replication, there are still open questions regarding how the proofreading exonuclease (NSP10/NSP14 complex) contributes to replication fidelity and resistance to nucleoside analogs. Through single turnover kinetic analysis, we show that the preferred substrate for the exonuclease is double-stranded RNA without any mismatches. Double-stranded RNA containing a 3′-terminal remdesivir was hydrolyzed at a rate similar to a correctly base-paired cognate nucleotide. Surprisingly, single-stranded RNA or duplex RNA containing a 3′-terminal mismatch was hydrolyzed at rates 125- and 45-fold slower, respectively, compared to the correctly base-paired double-stranded RNA. These results define the substrate specificity and rate of removal of remdesivir for the exonuclease and outline rigorous kinetic assays that could help in finding next-generation exonuclease inhibitors or nucleoside analogs that are able to evade excision. These results also raise important questions about the role of the polymerase/exonuclease complex in proofreading during viral replication. Addressing these questions through rigorous kinetic analysis will facilitate the search for desperately needed antiviral drugs to combat COVID-19.
Keywords: kinetics, proofreading exonuclease, SARS-CoV-2 polymerase, NSP14, remdesivir
Introduction
COVID-19, caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), presents a major global health threat that is unlikely to dissipate anytime soon. Scientists around the world have published enzymatic mechanisms and structures of viral proteins at an unprecedented rate. In particular, the kinetics and mechanism of fast RNA replication (300 nt/s) by the RNA-dependent-RNA polymerase (RdRp) and its inhibition by the nucleoside analog remdesivir have been established through a combination of kinetic and structural analyses.1,2 The bifunctional NSP10/NSP14 exonuclease/methyltransferase complex has been implicated in genome proofreading3,4 and RNA capping,5 and structural biologists have provided atomic structures of the exonuclease complex in isolation6−8 as well as bound to the RdRp complex (NSP12/NSP7/NSP8) and the NSP13 helicase.9 However, important details regarding kinetics and specificity of the proofreading exonuclease are lacking. Other groups have reported expression, purification, and preliminary activity assays for the SARS-CoV-2 exonuclease complex.3,6−8,10,11 From these studies, it is clear that the exonuclease activity of NSP14 (1) requires either Mg2+ or Mn2+ for catalysis4,6,7,10 (2) strongly prefers RNA bases over DNA bases,3,8 and (3) is greatly stimulated by the addition of the noncatalytic NSP10.11 However, there are contradictions in the literature as to whether a remdesivir incorporated into the primer strand of the RNA can be efficiently excised by the exonuclease complex7,8,12 and whether the enzyme preferentially hydrolyzes single-stranded RNA over base-paired RNA or RNA with terminal mismatches.3,4,7,8,10
To date, the ambiguity present in currently available studies on the exonuclease arises largely because initial experiments to characterize the exonuclease reaction have been performed under steady-state conditions with a single long time point, anywhere from 20 to 45 min, which does not provide a valid measure of substrate specificity.13 Fixed time point assays famously underestimate enzyme discrimination because the amounts of product formed after long incubation times do not reflect the different rates of reaction for various substrates. For example, while the preferred substrate may complete the reaction in a second, incubation for 30 min gives the alternative substrate nearly 2000-fold more time to complete the reaction. In the past, this error in experimental design has led to extraordinary misinterpretations, such as the conclusion that hydrogen bonds are not needed for efficient DNA replication, an error that was corrected by subsequent single turnover kinetic analysis.14 Moreover, steady-state assays using enzymes that operate on DNA are often rate-limited by the slow release of DNA from the enzyme, so they fail to measure the rates of catalysis and thereby greatly underestimate specificity.13 Single turnover kinetic experiments directly measure intrinsic rates of DNA cleavage or polymerization and can identify rate-limiting and specificity-determining steps in the reaction pathway that are masked by steady-state methods.1,13,15 Here, we present our initial work to characterize the substrate specificity and kinetics of the exonuclease hydrolysis reaction catalyzed by the NSP10/14 complex using transient kinetic methods. We performed experiments on three physiologically relevant substrates as well as an RNA substrate containing a remdesivir monophosphate incorporated into the primer strand. Our findings have implications for the design of experiments to screen viral exonuclease inhibitors and for the biological role of NSP10/14 beyond the proposed genome proofreading function.
Results
NSP10/NSP14 Exonuclease Hydrolyzes Mismatched and Single-Stranded RNA Much More Slowly Than Correctly Base-Paired RNA
We first measured rates of excision of the 3′-terminal nucleotide using 5′-[6-FAM] labeled RNA substrates using oligonucleotides based on those we previously used to measure kinetics of polymerization by the viral RdRp complex.1 The FAM label on the 5′-end of the primer strand was deemed to be outside of the oligonucleotide binding domain so that it would not interfere with binding at the active site, as shown in studies on DNA polymerases.15,16 As shown in Figure 1, we chose three physiologically relevant substrates to measure the rates of hydrolysis: single-stranded RNA (FAM-LS2), correctly base-paired primer/template double-stranded RNA (FAM-LS2/LS1), and primer/template double-stranded RNA containing a terminal A:A mismatch (FAM-LS2/LS1.2.2). Double-stranded RNA containing either correctly base-paired or mispaired 3′-terminal primer strands represents RNA replication intermediates that would be continuously formed by the RdRp during genome replication and possibly excised by the exonuclease. For well-characterized proofreading exonucleases, single-stranded nucleic acids are the preferred substrates.17,18 When primer extension at the polymerase active site stalls, the primer strand of duplex DNA can melt away from the complementary template strand and enter the exonuclease active site where the 3′-terminal base is rapidly excised.19
Figure 1.
Duplex RNA without mismatches is the preferred substrate for the SARS-CoV-2 NSP10/NSP14 exonuclease complex. A solution of 1 μM NSP10/NSP14 was mixed with 100 nM FAM-ssRNA (FAM-LS2, blue), FAM-primer/template double-strand RNA (dsRNA) with an A:A mismatch (FAM-LS2/LS1.2.2, green), or FAM-primer/template dsRNA (FAM-LS2/LS1, red) to start the reaction. Data are shown on a logarithmic time scale, fit to single-exponential functions, with observed rates of 0.016, 0.044, and 2 s–1 for ssRNA, A:A mismatch dsRNA, and primer/template dsRNA, respectively, as summarized in Table 1. Note that the template strands LS1 and LS1.2.2 differ by three nucleotides in the single-strand region of the template strand, which is unlikely to have any nearest-neighbor effects, which are due to duplex stability and structure.21
In our experiments, a solution of 1 μM NSP10/NSP14 complex was mixed with 100 nM FAM-labeled RNA in the presence of 5 mM Mg2+. The reaction was then stopped at various time points by quenching with EDTA, and products were resolved and quantified by capillary electrophoresis.20 The experiments were designed with a concentration of enzyme in large excess over the substrate so that complete substrate conversion to product could occur in a single turnover. In Figure 1, we show the loss of the full-length starting material over time for each RNA substrate, displayed on a logarithmic time scale and fit to a single-exponential function. Surprisingly, hydrolysis was fastest for the double-stranded RNA substrate without any mismatches (2 s–1), while hydrolysis of the double-stranded substrate with a mismatch and single-stranded RNA was 45- and 125-fold slower, respectively.
Since the primer/template RNA without mismatches was the preferred substrate, we performed further experiments to characterize the kinetics of this reaction. To determine whether the rate of RNA binding limited the observed rate of hydrolysis of the RNA substrate, we performed a rapid quench experiment with the dsRNA substrate using two protocols (Figure 2). In the first experiment, duplex RNA was mixed with the NSP10/14 exonuclease complex to start the reaction, as described above. Here, the enzyme must first bind to the RNA before hydrolyzing it, so the observed rate could be limited by the rate of RNA binding to the enzyme. In a separate experiment, we preincubated the RNA and the NSP10/14 complex in the presence of EDTA for 30 min before adding excess Mg2+ to start the reaction. In this experiment, the RNA equilibrates with the exonuclease complex during the preincubation step and catalysis is initiated upon adding the metal ions. For the experiment where RNA is mixed with the enzyme to start the reaction, the observed rate was 2 s–1, whereas the observed rate after adding Mg2+ to a preformed enzyme–RNA complex was approximately 5.4 s–1 (determined from the fit to the loss of the 20 nt starting material). These results suggest that the intrinsic rate of chemistry (cleavage) is at least 5.4 s–1 and the slower rate observed upon mixing enzyme with RNA may be limited by the rate of RNA binding. We observed a linear increase in the observed rate of RNA cleavage with increasing enzyme concentration (Figure S1) with an approximate apparent second-order rate constant of ∼2.2 μM–1 s–1, supporting a rate-limiting RNA binding step when the reaction was initiated by mixing RNA with 1 μM enzyme.
Figure 2.
Processive exonuclease kinetics observed with double-stranded RNA. (A) Excision upon mixing the enzyme and RNA to start the reaction. A solution of 1 μM NSP10/NSP14 complex was mixed with 100 nM FAM-LS2/LS1 RNA to start the reaction, monitored using a quench-flow instrument. Both syringes contained 5 mM Mg2+. Data for the loss of the 20 nt primer fit a single exponential with an observed rate of 2 ± 0.1 s–1. Data for the formation and decay of the 19 nt product best fit a double-exponential function with both rates at approximately 2 s–1, given in Table 1. (B) Excision upon the Mg2+ addition to E-RNA complex. A solution of 1 μM NSP10/NSP14, 2.5 mM EDTA, and 100 nM FAM-LS2/LS1 RNA was mixed with 7.5 mM Mg2+ to start the reaction. Samples were quenched with EDTA, and products were resolved by capillary electrophoresis. Fitting the fast phase of the loss of 20 nt starting material gave an observed rate of 6.5 ± 0.9 s–1. A double-exponential fit of the data for the formation and decay of the 19 nt intermediate is summarized in Table 1. Note that on the time scale of the experiment in panel (B) a fraction of the starting material (∼10%) fails to react. We could not account for this fraction of slow-reacting RNA by any simple model based on RNA–enzyme equilibration, so we have focused on the faster reaction phase accounting for 90% of the reaction.
We monitored the loss of the 20 nt starting material, the rise and fall of the 19 nt product, and the subsequent formation of products less than or equal to 18 nt in length (Figure 2) since peaks for the 17 and 18 nt product were not well resolved (as shown in Figures S2 and S3). Although the errors are somewhat larger for the double-exponential fit to define the rise and fall of the 19 nt intermediate, the comparable rates of formation and decay (Table 1), and the fact that the decay of the 19 nt intermediate goes to completion suggest that the enzyme does not dissociate after hydrolyzing one base but rather removes at least 2 nt in a processive manner. Similar experiments were performed with the ssRNA substrate and A:A mismatch dsRNA substrate, as shown in Figures S4–S7. For the ssRNA substrate (Figures S4 and S5), the observed rate of cleavage was comparable with or without preincubation to form the E–ssRNA complex. These results could suggest either weak binding of the RNA to the enzyme at the concentrations tested or a slow rate of chemistry. Structures of the complex showing double-stranded RNA stably bound at the exonuclease active site suggest that ssRNA may bind weakly.7 Data for the mismatched RNA substrate were biphasic upon the Mg2+ addition to the ES complex, with approximately half of the amplitude corresponding to the fast phase at a rate greater than 1 s–1 and the other half at an observed rate of 0.06 s–1 (Figure S7). These results suggest that for a mismatch, the intrinsic rate of cleavage may be relatively fast; however, the initial binding may be weak and limit the enzyme’s ability to process these substrates.
Table 1. Observed Ratesa.
| figure | substrate/phase | observed rate (s–1) | |
|---|---|---|---|
| Figure 1 | ssRNA (FAM-LS2) | 0.016 ± 0.0005 | |
| dsRNA—0 mismatches (FAM-LS2/LS1) | 2.0 ± 0.05 | ||
| dsRNA—A:A mismatch (FAM-LS2/LS1.2.2) | 0.046 ± 0.0033 | ||
| Figure 2 | RNA initiated reaction (A)—19 nt product | phase 1 | 2.3 ± 0.2 |
| phase 2 | 2.3 ± 0.2 | ||
| Mg2+ initiated reaction (B)—19 nt product | phase 1 | 6.5 ± 0.9 | |
| phase 2 | 5.4 ± 0.6 | ||
| Figure 3 | RMP RNA | fast phase | 2.4 ± 0.4 |
| slow phase | 0.42 ± 0.083 | ||
Here, we summarize the results of rate measurements. For the data in Figure 2, we show the rate measurements for the formation and decay of the 19 nt intermediate derived from a double-exponential fit. Fitting the fast phase for the loss of the starting 20 nt RNA gave an observed rate of 6.5 ± 0.9 s–1. In fitting these data, we constrained the fast phase for the loss of 20 nt and formation of 19 nt intermediate to follow the same observed rate.
Remdesivir Is Hydrolyzed at a Rate Comparable to Correctly Base-Paired RNA
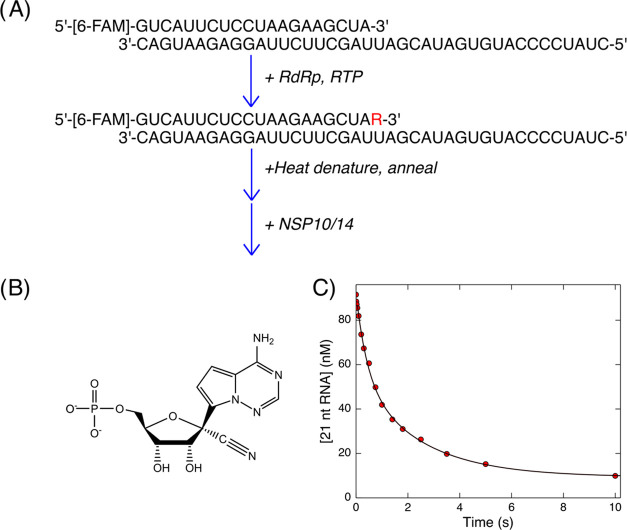
Remdesivir is a nucleoside analog drug that, in its triphosphate form (RTP), is rapidly incorporated by the SARS-CoV-2 RdRp.1 Remdesivir was approved by the FDA after it was shown to be effective in treating COVID-19, provided the drug is given early in the course of infection.22 Others have shown that a remdesivir monophosphate incorporated into a primer strand can be hydrolyzed8 contrary to earlier predictions that this nucleoside analog would resist exonuclease hydrolysis due to the 1′ cyano group.12 The rates of this hydrolysis remain to be quantitatively measured to estimate the effect of the 1′ cyano group. We therefore directly measured this rate of hydrolysis using our pre-steady-state exonuclease assay. To address whether remdesivir monophosphate (RMP) incorporated into an RNA primer strand is efficiently hydrolyzed, we used the purified SARS-CoV-2 RdRp complex to enzymatically synthesize an RNA substrate containing a terminal RMP (see the Materials and Methods section). We achieved a 92% efficiency in synthesizing the RNA primer containing RMP, as quantified by capillary electrophoresis. We then heat-denatured the RdRp complex and reannealed the primer/template duplex RNA. We performed the excision reaction on the resulting RNA in the quench flow by mixing with 1 μM NSP10/NSP14 complex and analyzed the time course of RNA cleavage by capillary electrophoresis. The reaction is processive, as shown in Figure S8. A fit to the time dependence of RMP removal is shown in Figure 3. The data best fit a double-exponential function (for comparison, a single-exponential fit is shown in Figure S9) with similar amplitudes for the fast and slow phases with observed rates of 2.4 and 0.42 s–1, respectively. The reason for two phases of the reaction is still unknown but could represent two different binding modes of the terminal RNA to the enzyme or different conformations of the remdesivir in the RNA. Nonetheless, these results show that at least half of the terminal remdesivir is hydrolyzed at a rate comparable to correctly base-paired RNA, showing that it is easily removed after incorporation into the genome. Nonetheless, it is likely that remdesivir could escape removal by the proofreading exonuclease since the RNA primer is extended more rapidly than RMP is excised and polymerization stalls only after the incorporation of three additional nucleotides on top of RMP.1,2
Figure 3.
Excision of incorporated remdesivir monophosphate. (A) Scheme for the enzymatic synthesis of RMP containing substrate. SARS-CoV-2 RdRp complex and remdesivir triphosphate were added to enzymatically incorporate RMP into the primer strand (see the Materials and Methods section). The RdRp complex was heat-denatured, and then the RNA was reannealed before measuring excision by NSP10/NSP14 in the quench flow. (B) Structure of remdesivir monophosphate. Minor modifications relative to ATP in the ring and the addition of a 1′ cyano group that causes delayed chain termination by the RdRp.1,2 (C) Time course of remdesivir excision from the RNA. Data are shown fit to a double-exponential function with rates of 2.4 and 0.42 s–1 for the fast and slow phases, respectively. Amplitudes for the two phases are approximately equal.
Discussion
Steady-state experiments commonly performed on the NSP10/NSP14 exonuclease are difficult to interpret since the exonuclease reaction is over in a fraction of a second, while steady-state time scales close to 1 h are typically employed. Single turnover experiments have the advantage of directly measuring the relative rates of excision on biologically relevant time scales with enzyme in excess to monitor reactions occurring at the active site of the enzyme. These methods provide quantitative results that can be directly interpreted and include estimates of the concentration of active enzyme. The ambiguity of the steady-state results and the lack of a standard for acceptable enzyme activity in the literature provided the motivation for the direct experiments outlined in this paper to unambiguously address these questions.
We began by choosing three RNA substrates on which to measure excision. Most proofreading exonucleases function on at least partially single-stranded nucleic acids23 arising from the melting of duplex DNA or RNA at the polymerase active site to transfer the 3′-end of the primer into the exonuclease active site.17,19,24−26 Our data show that, unlike other proofreading exonucleases, neither single-stranded RNA nor double-stranded RNA containing a mismatch is a preferred substrate, exhibiting rates of hydrolysis 45- to 125-fold slower than rates of excision on correctly base-paired RNA. This observation may imply that the RNA duplex must dissociate completely from the polymerase active site and both strands of the RNA must rebind at the exonuclease active site without tethering the template strand in the polymerase active site, as previously suggested.9 While the kinetics for the NSP10/NSP14 complex in isolation is clear, the major limitation of this study and others is that the experiments fail to consider that the exonuclease presumably functions in a complex containing the helicase, RdRp, and potentially other viral nonstructural proteins, which may change the kinetics of RNA hydrolysis and substrate specificity. For example, a recent preprint suggests that the kinetics of exonuclease hydrolysis changes in the presence of NSP16.27 Previous studies have clearly shown a robust change in activity upon the association of NSP10 with NSP14, so a further change in activity when complexed with other viral proteins is conceivable. High-quality single turnover kinetic analysis of the exonuclease complex in conjunction with the RdRp and other viral nonstructural proteins are required to address these questions about whether the substrate specificity of the NSP10/14 complex changes in conjunction with other binding partners. Moreover, the critical role of the proofreading exonuclease will rely on the relative rates of primer extension versus excision from a single multienzyme complex containing both activities, as shown for DNA polymerases.17,19
We also quantified rates of excision of remdesivir monophosphate incorporated into the primer strand of a duplex RNA and found that this analog was hydrolyzed at a rate similar to that for correctly base-paired RNA. While remdesivir is incorporated with higher efficiency than ATP by the viral RdRp,1 the high excision efficiency we observed here is slower than the rate of extension. Thus, remdesivir may be protected from the proofreading exonuclease by being rapidly buried by subsequent incorporation of normal nucleotides. By stalling polymerization only after being buried by three nucleotides, remdesivir would be protected from immediate removal by the exonuclease.1,2 Nonetheless, this protection is not absolute, and processive exonuclease activity could ultimately remove RMP. Further studies are currently underway to measure the kinetics of excision of molnupiravir, another promising nucleoside analog.28
While to the best of our knowledge no one has designed a nucleoside analog that evades excision by the exonuclease, it is possible that certain modifications could inhibit exonuclease hydrolysis. If such modifications were also efficiently incorporated by the polymerase, such a molecule would presumably be an attractive candidate for antiviral studies in vivo. Alternatively, exonuclease inhibitors, which have been successfully discovered in vitro,29 could be used in combination with nucleoside analog therapies to make both inhibitors more potent. Single turnover experiments using the methods outlined in this paper and our previous studies could prevent many of the pitfalls of steady-state experiments on these enzymes and provide direct, quantitative answers on the potential effectiveness of new inhibitors.
Besides proofreading, the exonuclease activity of the NSP10/14 complex may have other roles in the life cycle of the virus. One study proposed a role in the translational shutdown of the infected host cell,30 although the mechanism of this remains unknown. When exonuclease activity is knocked out in cell culture experiments, SARS-CoV-2 curiously fails to replicate rather than simply accumulating more mutations31 as would be expected if the exonuclease activity only functions in proofreading. Interestingly, for reasons that are still unknown, this is not the case for the original SARS-CoV.32 One role of exonuclease activity that has been proposed is in viral recombination,31 which is consistent with our data where the enzyme would efficiently hydrolyze stretches of correctly base-paired RNA on different viral genomic templates allowing for efficient recombination. Efficient recombination is a hallmark of coronaviruses and could contribute to the large number of variants that have evolved relatively quickly in the COVID-19 pandemic. The necessity of the exonuclease complex for virus propagation makes this complex an attractive target for the design of next-generation exonuclease inhibitors that may boost the efficacy of currently available antiviral drugs. Studies outlined here provide a boilerplate for the analysis and evaluation of other nucleotide analogs designed to inhibit SARS-CoV-2 RNA replication.
Materials and Methods
Enzymes, Oligonucleotides, and Reagents
Expression and Purification of NSP10/14
NSP10 and NSP14 were expressed and purified as previously described.29 Briefly, the coexpression of NSP10/14 with N-terminal Strep tag II and N-terminal 8xHis tag, respectively, in BL21(DE3) E. coli was induced with 0.5 mM IPTG at 16 °C for 20 h. Cells were harvested, lysed, then purified on a low-resolution Ni-NTA column (Qiagen), dialyzed, and purified on a high-resolution Ni-NTA column (HiTrap IMAC HP, GE). The complex was then further purified by size exclusion chromatography (Superdex 200, GE), flash-frozen, and stored at −80 °C.
Expression and Purification of NSP12-His/NSP7L8
His-tagged NSP12 and NSP7L8 were coexpressed and purified as previously described.1 Briefly, the coexpression of 8xC-His-tagged NSP12 and NSP7L8 in BL21 E. coli/pG-Tf2 was induced with 0.5 mM IPTG, 10 ng/mL tetracycline, and 50 μg/mL nalidixic acid at 16 °C for 16 h. Harvested cells were lysed, clarified by centrifugation, and purified by Ni-NTA chromatography (His-Trap FF, Cytiva). Fractions containing the NSP12-His/NSP7L8 complex were flash-frozen in liquid nitrogen and stored at −80 °C.
Oligonucleotides and Reagents
RNA oligonucleotides were synthesized by Integrated DNA Technologies with RNase-free HPLC purification. The concentration of purified oligonucleotides was determined by absorbance at 260 nm using the extinction coefficients given in Table 2. Oligo stocks were stored in annealing buffer (10 mM tris–HCl pH 7, 50 mM NaCl, 0.1 mM EDTA) at −20 °C. Double-stranded RNA substrates were prepared by mixing the primer and template strands at a 1:1 molar ratio in annealing buffer, heating to 90 °C for 3 min, and then cooling slowly to room temperature over the course of 1 h. Remdesivir triphosphate (GS-443902, RTP) was kindly provided by Joy Feng and Brian Schultz of Gilead Sciences. The concentration of RTP was determined by absorbance at 245 nm using an extinction coefficient of 24,100 M–1 cm–1.1 All buffer components and other chemicals were purchased from either Sigma-Aldrich or Thermo Fisher Scientific.
Table 2. Oligonucleotides Used in This Study.
| oligo name | sequence 5′–3′ | extinction coefficient at 260 nm (M–1 cm–1) |
|---|---|---|
| FAM-LS2 | [6-FAM]—GUCAUUCUCCUAAGAAGCUA | 222,360 |
| LS1 | CUAUCCCCAUGUGAUUUUAAUAGCUUCUUAGGAGAAUGAC | 403,100 |
| LS1.2-U | CUAUCCCCAUGUGAUACGAUUAGCUUCUUAGGAGAAUGAC | 401,600 |
| LS1.2.2 | CUAUCCCCAUGUGAUACGAAAAGCUUCUUAGGAGAAUGAC | 404,600 |
| Cy3–28mer DNA | [Cy3]-CCGTGAGTTGGTTGGACGGCTGCGAGGC | 266,800 |
Enzymatic Synthesis of Remdesivir-Containing RNA Substrate
An RNA substrate containing a terminal remdesivir monophosphate was synthesized enzymatically using the following protocol. A solution of 2.5 μM NSP12-His/NSP7L8,1 5 μM NSP8, 200 nM FAM-LS2/LS1.2-U RNA, and 20 μM RTP was incubated for 90 min at room temperature to form approximately 92% product RNA containing a terminal remdesivir monophosphate. The reaction was heated to 95 °C for 5 min, cooled slowly to room temperature over the course of 1 h, and then centrifuged at 20,000g in a tabletop microcentrifuge for 20 min to pellet the precipitated protein. The supernatant was then used in kinetics experiments.
Kinetics Experiments
All kinetics experiments were performed at 37 °C in SARS-CoV-2 reaction buffer (40 mM tris–HCl pH 7, 50 mM NaCl, 5 mM MgCl2, 1 mM DTT).1,2 In some experiments, Mg2+ was only present in one syringe but at 2× the final concentration to measure the kinetics of hydrolysis of RNA already bound to the enzyme. Rapid quench experiments were performed using a KinTek RQF-3 (KinTek Corp) with reaction buffer in the drive syringes and 0.6 M EDTA in the quench syringe for a final concentration of 0.2 M after quenching. A circulating water bath was used for temperature control. Kinetics time points were analyzed by capillary electrophoresis on an ABI 3130xl Genetic Analyzer with a 36 cm array and a nanoPOP-6 polymer (Molecular Cloning Laboratories) at 65 °C. Samples were prepared for analysis by mixing 1 μL of sample with 10 μL of HiDi formamide (Thermo Fisher) containing a 28 nt Cy3-labeled DNA oligo internal standard for sizing. Samples were injected for 6–12 s, depending on the experiment, at 3.6 kV. The peak area was determined with GeneMapper software, and sizing and quantification were performed with a program written in house.2 Concentrations of reaction components given in the text are final concentrations after mixing unless otherwise noted. Experiments were repeated at least once to ensure reproducibility. Electropherograms from which figures in the main paper are derived are given in the Supporting Information.
Data Fitting and Analysis
Data fitting and analysis were performed using KinTek Explorer simulation and data fitting software v11 (www.kintekexplorer.com).33,34 This software was also used in preparing figures for kinetic data. Conventional data fitting was performed in the software using built-in functions. The equation for a single exponential is y = A0 + A1(1 – exp(−b1t)), where A0 is the y-value at time zero, A1 is the amplitude, b1 is the decay rate, and t is time. The equation for a double exponential is y = A0 + A1(1 – exp(−b1t)) + A2(1 – exp(−b2t)), where A0 is the y-value at time zero, A1 and A2 are the amplitudes of the first and second phases, b1 and b2 are the decay rates of the first and second phases, respectively, and t is time.
Acknowledgments
The authors would like to thank Brian Schultz and Joy Feng at Gilead Sciences for kindly providing the remdesivir triphosphate used in this study and Jerome Deval and Cheng Liu at Aligos therapeutics for providing the purified NSP10/NSP14 used in this study, expressed and purified as described.29
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsbiomedchemau.2c00046.
Data showing the enzyme concentration dependence of cleavage; electropherograms showing raw data for experiments described in the main text; global data fitting defining the kinetics of processive exonuclease reactions; and alternative (single-exponential) fit of remdesivir excision data (PDF)
Author Contributions
CRediT: Tyler L. Dangerfield conceptualization, formal analysis, investigation (lead), methodology (lead), visualization (lead), writing-original draft (lead), writing-review & editing (equal); Kenneth A Johnson conceptualization (equal), formal analysis (equal), funding acquisition (lead), project administration (lead), software (lead), supervision (lead), writing-review & editing (equal).
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases (NIAID) to K.A.J. (NIH R01AI163336).
The authors declare the following competing financial interest(s): K.A.J. is president of KinTek Corporation, which provided the RQF-3 rapid quench flow instrument and KinTek Explorer software used in this study.
Supplementary Material
References
- Dangerfield T. L.; Huang N. Z.; Johnson K. A. Remdesivir Is Effective in Combating COVID-19 because It Is a Better Substrate than ATP for the Viral RNA-Dependent RNA Polymerase. iScience 2020, 23, 101849 10.1016/j.isci.2020.101849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J. P. K.; Dangerfield T. L.; Taylor D. W.; Johnson K. A. Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication. Mol. Cell 2021, 81, 1548–1552.e4. 10.1016/j.molcel.2021.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddock H. T.; Brolih S.; Yosaatmadja Y.; Ratnaweera M.; Bielinski M.; Swift; Lonnie P.; Cruz-Migoni A.; Fan H.; Keown J. R.; Walker A. P.; et al. Characterization of the SARS-CoV-2 ExoN (nsp14ExoN–nsp10) complex: implications for its role in viral genome stability and inhibitor identification. Nucleic Acids Res. 2022, 50, 1484–1500. 10.1093/nar/gkab1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier M. N.; Riccio A. A.; Wilson I. M.; Copeland W. C.; Stanley R. E. Recent insights into the structure and function of coronavirus ribonucleases. FEBS Open Bio 2022, 12, 1567–1583. 10.1002/2211-5463.13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M.; Debarnot C.; Imbert I.; Selisko B.; Snijder E. J.; Canard B.; Decroly E. In Vitro Reconstitution of SARS-Coronavirus mRNA Cap Methylation. PLoS Pathog. 2010, 6, e1000863 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.; Chen H.; Chen Z.; Yang F.; Ye F.; Zheng Y.; Yang J.; Lin X.; Sun H.; Wang L.; et al. Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity. Nucleic Acids Res. 2021, 49, 5382–5392. 10.1093/nar/gkab320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.; Shi W.; Becker S. T.; Schatz D. G.; Liu B.; Yang Y. Structural basis of mismatch recognition by a SARS-CoV-2 proofreading enzyme. Science 2021, 373, 1142–1146. 10.1126/science.abi9310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller N. H.; Shi K.; Demir Ö.; Belica C.; Banerjee S.; Yin L.; Durfee C.; Amaro R. E.; Aihara H. Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN. Proc. Natl. Acad. Sci. U.S.A. 2022, 119, e2106379119 10.1073/pnas.2106379119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L.; Yang Y.; Li M.; Zhang Y.; Zheng L.; Ge J.; Huang Y. C.; Liu Z.; Wang T.; Gao S.; et al. Coupling of N7-methyltransferase and 3′-5′ exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading. Cell 2021, 184, 3474–3485.e11. 10.1016/j.cell.2021.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z.; Pourfarjam Y.; Kim I. K. Reconstitution and functional characterization of SARS-CoV-2 proofreading complex. Protein Expression Purif. 2021, 185, 105894 10.1016/j.pep.2021.105894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio A. A.; Sullivan E. D.; Copeland W. C. Activation of the SARS-CoV-2 NSP14 3′-5′ exoribonuclease by NSP10 and response to antiviral inhibitors. J. Biol. Chem. 2022, 298, 101518 10.1016/j.jbc.2021.101518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon A.; Le N. T.; Selisko B.; Eydoux C.; Alvarez K.; Guillemot J. C.; Decroly E.; Peersen O.; Ferron F.; Canard B. Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antiviral Res. 2020, 178, 104793 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. A.Kinetic Analysis for the New Enzymology; KinTek Corporation, 2019. [Google Scholar]
- Lee H. R.; Helquist S. A.; Kool E. T.; Johnson K. A. Importance of hydrogen bonding for efficiency and specificity of the human mitochondrial DNA polymerase. J. Biol. Chem. 2008, 283, 14402–14410. 10.1074/jbc.M705007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangerfield T. L.; Johnson K. A. Conformational dynamics during high-fidelity DNA replication and translocation defined using a DNA polymerase with a fluorescent artificial amino acid. J. Biol. Chem. 2021, 296, 100143 10.1074/jbc.RA120.016617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanes J. W.; Johnson K. A. Analysis of single nucleotide incorporation reactions by capillary electrophoresis. Anal. Biochem. 2005, 340, 35–40. 10.1016/j.ab.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Donlin M. J.; Patel S. S.; Johnson K. A. Kinetic partitioning between the exonuclease and polymerase sites in DNA error correction. Biochemistry 1991, 30, 538–546. 10.1021/bi00216a031. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz L. J.; Marquez L. A.; Elisseeva E.; Baker R. P.; Bloom L. B.; Dunford H. B.; Goodman M. F. The Proofreading Pathway of Bacteriophage T4 DNA Polymerase. J. Biol. Chem. 1998, 273, 22969–22976. 10.1074/jbc.273.36.22969. [DOI] [PubMed] [Google Scholar]
- Dangerfield T. L.; Kirmizialtin S.; Johnson K. A. Substrate specificity and proposed structure of the proofreading complex of T7 DNA polymerase. J. Biol. Chem. 2022, 298, 101627 10.1016/j.jbc.2022.101627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangerfield T. L.; Huang N. Z.; Johnson K. A. High throughput quantification of short nucleic acid samples by capillary electrophoresis with automated data processing. Anal. Biochem. 2021, 629, 114239 10.1016/j.ab.2021.114239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee D.; Tateishi-Karimata H.; Ohyama T.; Ghosh S.; Endoh T.; Takahashi S.; Sugimoto N. Improved nearest-neighbor parameters for the stability of RNA/DNA hybrids under a physiological condition. Nucleic Acids Res. 2020, 48, 12042–12054. 10.1093/nar/gkaa572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo P.; Tacconelli E.; Olimpieri P. P.; Celant S.; Colatrella A.; Tomassini L.; Palù G. Mortality in SARS-CoV-2 Hospitalized Patients Treated with Remdesivir: A Nationwide, Registry-Based Study in Italy. Viruses 2022, 14, 1197 10.3390/v14061197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont P. S.; Friedman J. M.; Beese L. S.; Sanderson M. R.; Steitz T. A. Cocrystal Structure of an Editing Complex of Klenow Fragment with DNA. Proc. Natl. Acad. Sci. U.S.A. 1988, 85, 8924–8928. 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd T.; Botto M.; Paul F.; Fernandez-Leiro R.; Lamers Meindert H.; Ivanov I. Polymerization and editing modes of a high-fidelity DNA polymerase are linked by a well-defined path. Nat. Commun. 2020, 11, 5379 10.1038/s41467-020-19165-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane R.; Berezhna S. Y.; Gill J. P.; Van der Schans E.; Millar D. P. Dynamics of Site Switching in DNA Polymerase. J. Am. Chem. Soc. 2013, 135, 4735–4742. 10.1021/ja311641b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman K. R.; Dahl J. M.; Wang H. Kinetic Mechanism at the Branchpoint between the DNA Synthesis and Editing Pathways in Individual DNA Polymerase Complexes. J. Am. Chem. Soc. 2014, 136, 7117–7131. 10.1021/ja5026408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda A.; Plewka J.; Chykunova Y.; Jones A. N.; Pachota M.; Rawski M.; Mourão A.; Karim A.; Kresik L.; Lis K.; et al. Despite the odds: formation of the SARS-CoV-2 methylation complex. Submitted 2022-03-01. bioRxiv https://www.biorxiv.org/content/10.1101/2022.01.25.477673v2 (accessed 2022-07-01). [DOI] [PMC free article] [PubMed]
- Kabinger F.; Stiller C.; Schmitzova J.; Dienemann C.; Kokic G.; Hillen H. S.; Hobartner C.; Cramer P. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021, 28, 740–746. 10.1038/s41594-021-00651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholle M. D.; Liu C.; Deval J.; Gurard-Levin Z. A. Label-Free Screening of SARS-CoV-2 NSP14 Exonuclease Activity Using SAMDI Mass Spectrometry. SLAS Discovery 2021, 26, 766–774. 10.1177/24725552211008854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J. C.-C.; Laurent-Rolle M.; Pawlak J. B.; Wilen C. B.; Cresswell P. Translational shutdown and evasion of the innate immune response by SARS-CoV-2 NSP14 protein. Proc. Natl. Acad. Sci. U.S.A. 2021, 118, e2101161118 10.1073/pnas.2101161118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando N. S.; Zevenhoven-Dobbe J. C.; van der Meer Y.; Bredenbeek P. J.; Posthuma C. C.; Snijder E. J. The Enzymatic Activity of the nsp14 Exoribonuclease Is Critical for Replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020, 94, e01246-01220 10.1128/JVI.01246-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando N. S.; Ferron F.; Decroly E.; Canard B.; Posthuma C. C.; Snijder E. J. The Curious Case of the Nidovirus Exoribonuclease: Its Role in RNA Synthesis and Replication Fidelity. Front. Microbiol. 2019, 10, 1813 10.3389/fmicb.2019.01813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. A.; Simpson Z. B.; Blom T. Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal. Biochem. 2009, 387, 20–29. 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- Johnson K. A. Fitting enzyme kinetic data with KinTek global kinetic explorer. Methods Enzymol. 2009, 467, 601–626. 10.1016/S0076-6879(09)67023-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.