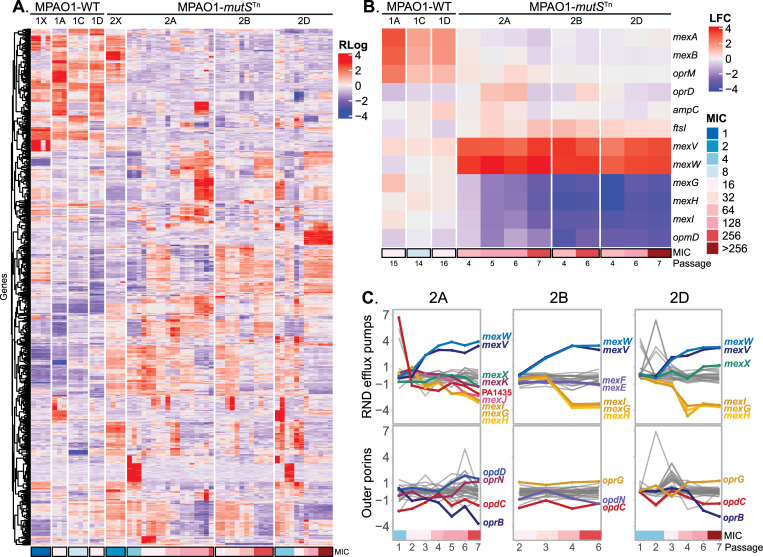
Fig 3. Dissimilar transcriptome profiles highlight lack of involvement of major cephalosporin resistance genes in CZA resistance in MMR-deficient lineages and demonstrate MexVW overexpression.
Three lineages of MPAO1-WT and MPAO1-mutSTn were selected for transcriptomic analysis and 3 biological replicates from 1 isolate per passage underwent RNA sequencing. (A) A clustered heatmap of the top 750 most variable genes across CZA-resistant isolates is shown. Rlog normalized levels of expression of each gene (rows) were scaled within each row. Isolates (columns) are grouped by lineage and arranged by genotype with WT on the left and hypermutators on the right. Within each lineage, isolates are arranged from left to right in order of increasing number of passages under CZA selection, with the corresponding MIC indicated at the bottom. 1X and 2X represent the MPAO1-WT and MPAO1-mutSTn parental strains of the respective lineages. (B) LFC with respect to their corresponding ancestor of the terminal MPAO1-WT isolates (first 3 columns) and CZA-adapted MPAO1-mutSTn isolates (last 9 columns). The rows represent 6 major beta-lactam resistance genes in P. aeruginosa from 4 major resistance pathways (encoding the MexAB-OprM efflux pump, OprD outer membrane porin, PDC beta-lactamase, and penicillin-binding-protein 3 FtsI). Data for 2 differentially expressed RND efflux pumps in the MPAO1-mutSTn lineages (MexVW and MexGHI-OpmD) are additionally shown. (C) Expression levels of RND efflux pumps and outer membrane porins in MPAO1-mutSTn lineages during the course of the CZA adaptation experiment. The vertical axis represents the LFC of expression levels compared to the early isolates, and the horizontal axis shows the passage number. CZA MIC is indicated with color tiles under the axis. Lines are colored and labeled if the respective gene reached a LFC > 1.0 in the terminal isolates of the lineage. The underlying data to generate this figure can be found in S2 Data. CZA, ceftazidime/avibactam; MMR, mismatch repair; WT, wild type.