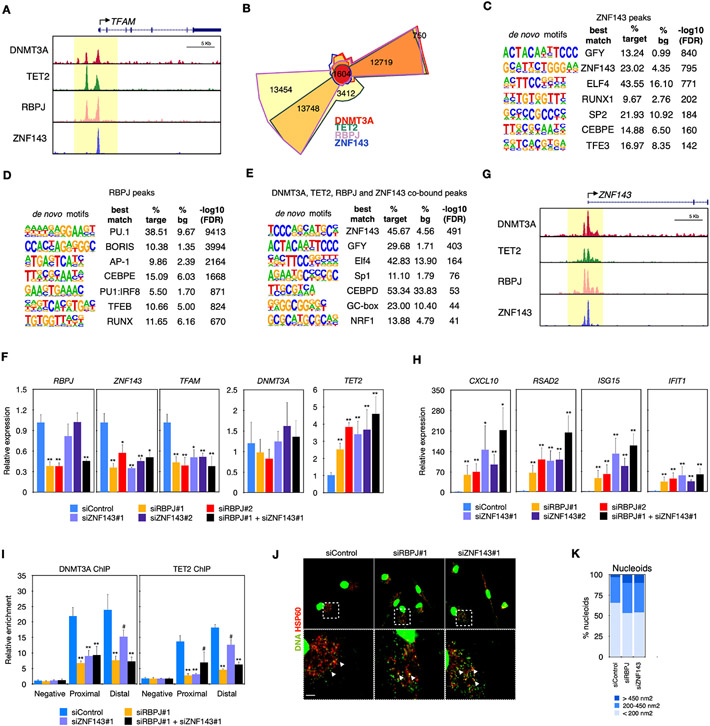
Figure 7. Combinatorial binding of RBPJ, ZNF143, DNMT3A, and TET2 to coordinate TFAM expression and prevent type-I IFN signaling.
(A) Composites of DNTM3A, TET2, RBPJ ,and ZNF143 ChIP-seq showing binding to TFAM promoter.
(B) Chow-Ruskey plot showing the overlap between DNTM3a, TET2, ZNF143, and RBPJ ChIP-seqs (n = 4 donors for DNMT3A and TET2 ChIP-seqs and n = 2 donors for ZNF143 and RBPJ ChIP-seq).
(C) HOMER analysis of motifs found in the 1,604 peaks co-bound by DNMT3A, TET2, RBPJ, and ZNF143 showing motifs for ZNF143, ELF4, PU.1, and CEBPD.
(D) HOMER analysis of motifs found in ZNF143 peaks showing enrichment for ZNF143, ELF4, RUNX, PU.1, CEBPE, and MITF/TFE.
(E) HOMER analysis of motifs found in RBPJ peaks showing enrichment in PU.1, BORIS, AP-1, CEBPE, PU.1-IRF8, and MITF/TFE.
(F) RT-qPCR of RBPJ, ZNF143, DNTM3A, TET2, or TFAM in MDM treated with siRNAs for RBPJ or ZNF143 (n = 3 donors).
(G) Composites of DNTM3A, TET2, RBPJ, and ZNF143 ChIP-seq showing binding to ZNF143 promoter.
(H) RT-qPCR showing upregulation of CXCL10, RSAD2, ISG15, and IFIT1 in MDM treated with siRNAs for RBPJ or ZNF143 (n = 3 donors).
(I) ChIP-qPCR over a control negative region (negative), proximal or distal region of TFAM promoter showing reduced binding of DNMT3A and TET2 in MDM treated with siRNAs for RBPJ or ZNF143 (n = 3 donors).
(J) Representative images by IF analysis of HSP60 and DNA showing larger nucleoids (arrowheads) in macrophages with DNMT3A or TET2 mutations. Scale bars, 2 μm.
(K) Quantification of cytosolic DNA showing increased number of macrophages treated with siRNA for RBPJ or ZNF143 or TFAM. Mann-Whitney U test was used to calculate statistical significance. *p < 0.05; **p < 0.01. Also see Figure S7.