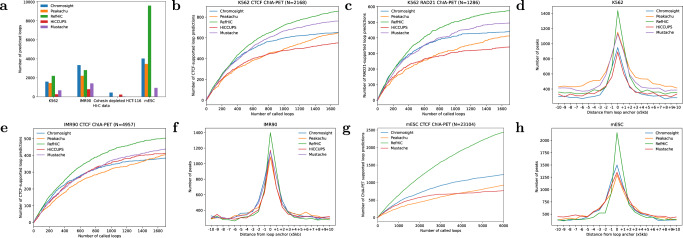
Fig. 3. Loop detection in Hi-C data from human K562, IMR90, and cohesin-depleted HCT-116 cells, as well as mouse ESC.
a Number of loops identified in Hi-C datasets obtained in each data set (human data: test chromosomes 15–17 only; mouse data: all autosomes). Note that in HCT-116, one would not expect any cohesin-mediated loops. b, c Number of ChIA-PET/HiCHIP-supported loop predictions, among the top predictions made by RefHiC and other tools on K562 Hi-C data, for test chromosomes chr15-17, compared against CTCF (b) and RAD21 ChIA-PET (c) data. d Occupancy of ChIP-seq identified CTCF binding sites in K562 cells as a function of distance to predicted loop anchors. e, f Same as (b, d), but for data obtained in IMR90 cells. g, h Same as (b, d), but for data obtained in mESC (all autosomes).