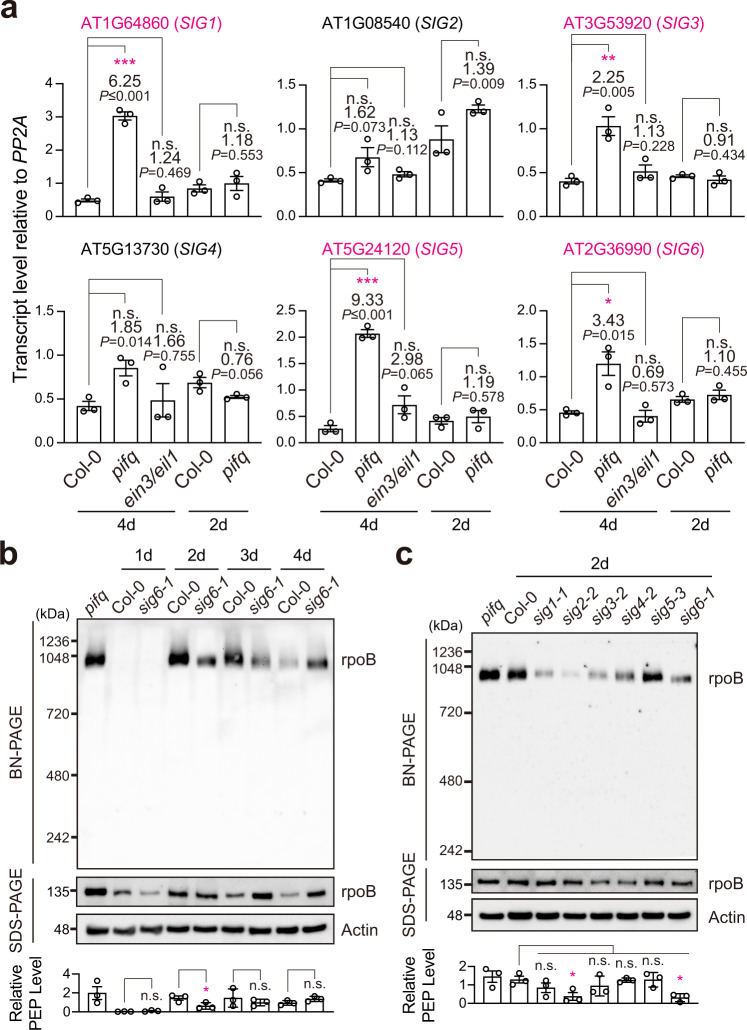
Fig. 7. PEP activation correlates with the expression of PIF-repressed sigma factors.
a qRT-PCR analysis of the steady-state transcript levels of SIG1-6 in dark-grown 4-d-old Col-0, pifq, and ein3/eil1, as well as 2-d-old Col-0 and pifq seedlings. The transcript levels were calculated relative to those of PP2A. The fold changes between the transcript levels in the mutants and their respective Col-0 controls are shown. Asterisks indicate statistically significant differences in the transcript level compared with that of Col-0 based on two-tailed Student’s t-test (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). If the change was less than two-fold or not statistically significant, it is labeled as n.s. (not significant). Error bars represent the s.e. of three biological replicates, and the centers of the error bars represent the mean values. b Immunoblots showing the levels of the PEP complex and the core PEP component rpoB in Col-0, and sig6-1 seedlings grown in 10 μmol m−2sec−1 R light for 1 to 4 days after imbibition. c Immunoblots showing the levels of the PEP complex and the core PEP component rpoB in 2-d-old Col-0, sig1-1, sig2-2, sig3-2, sig4-2, sig5-3, and sig6-1 seedlings grown in 10 μmol m−2sec−1 R light. For b and c, 4-d-old pifq seedlings grown in the dark were used as a control. Total protein was isolated under either native or denaturing conditions and resolved BN-PAGE or SDS-PAGE to assess the fraction of rpoB in the PEP complex or the amount of total rpoB by immunoblots, respectively. Actin was used as a loading control. The graph below the immunoblots shows the relative PEP levels, which were estimated using the relative level of rpoB in the PEP complex (BN-PAGE) divided by the relative level of denatured rpoB (SDS-PAGE) in each sample. Asterisks indicate statistically significant differences with Col-0 in triplicates based on two-tailed Student’s t-test (*P ≤ 0.05). Error bars represent the s.e. of three biological replicates, and the centers of the error bars represent the mean values. The source data underlying the qRT-PCR analysis in a and the immunoblots in b and c are provided in the Source Data file.