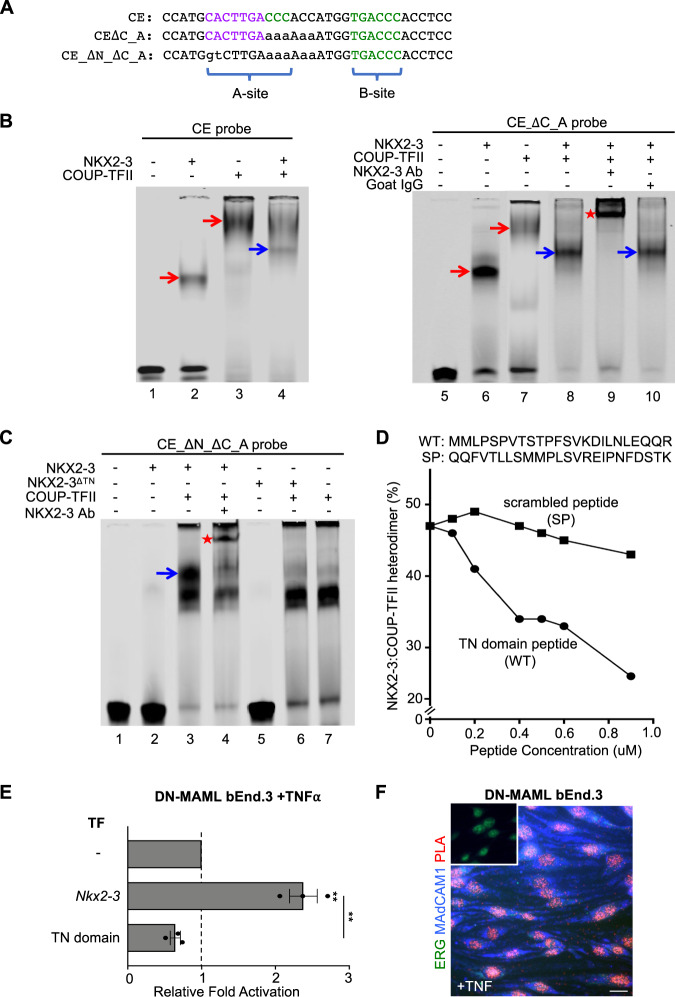
Fig. 2. NKX2-3 and COUP-TFII heterodimerize via the tinman (TN) domain to activate MAdCAM1.
A Sequence of wildtype and mutant CE probes for EMSA. B EMSA showing migration of recombinant NKX2-3 and COUP-TFII bound to the CE probe (left) and to CE_ΔC_A probe that lacks the COUP-TFII “A” site (right). Co-incubation of NKX2-3 and COUP-TFII yields a distinct band (blue arrows) reflecting an NKX2-3:COUP-TFII DNA complex that is supershifted by anti-NKX2-3 antibody but not by control antibody. C EMSA of protein complexes formed by incubation of the indicated TFs with a mutant CE probe lacking the NKX2-3 and the COUP-TFII “A” binding sites (CE_ΔN_ΔC_A). The probe binds COUP-TFII (lane 7) and not NKX2-3 (lane 2), but seeds a COUP-TFII:NKX2-3:DNA complex (blue arrow) when co-incubated with both TFs. The heterodimer-DNA complex is supershifted by anti-NKX2-3 antibody (red star). NKX2-3ΔTN failed to heterodimerize with COUP-TFII (lane 6). D Disruption of the NKX2-3:COUP-TFII heterodimer-DNA complex by a competing NKX2-3 TN domain peptide (WT) or by a scrambled peptide (SP). Data are expressed as percent of probe bound by NKX2-3:COUP-TFII heterodimer. E Madcam1 expression in TNFα-treated DN-MAML bEnd.3 cells stably overexpressing Nkx2-3 or the Nkx2-3 TN domain, as evaluated by real-time PCR. Values are mean ± SEM of three biological replicates. Two tailed t-test, paired. **: p value < 0.01. F Proximity ligation assay of NKX2-3 and COUP-TFII in TNFα-treated DN-MAML bEnd.3 cells stained for MAdCAM1 (blue) and ERG (green), with the proximity ligation signal shown in red. Inset shows a negative control with isotype-matched primary antibodies. Scale bar: 20m.