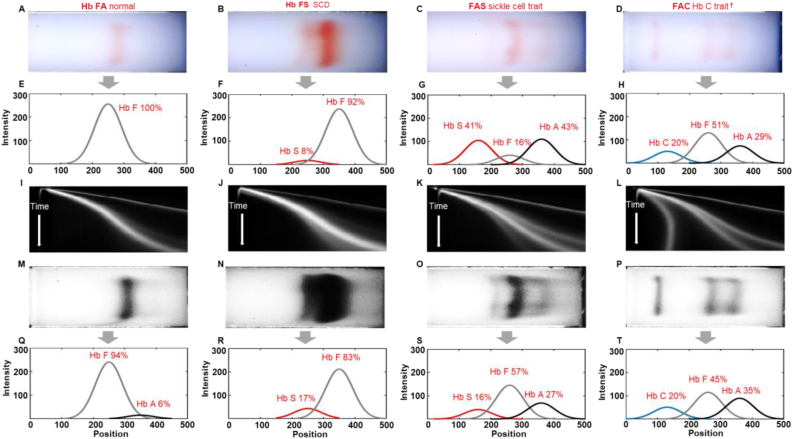
Figure 2.
Identification of Hb variants and quantification of Hb percentages by Gazelle-Multispectral. (A-D) The first row shows images captured under white light field. (E-H) The second row shows electropherograms reconstructed by the white data analysis algorithm based on the white light images (electropherograms not visible by users in the field). (I-L) The third row illustrates 2D representation of Gazelle-Multispectral space-time plots of band migration in 410 nm imaging mode, which are used with the machine learning algorithm for identifying Hb variants. (M-P) The fourth row shows images captured under 410 nm wavelength. (Q-T) The fifth row shows electropherograms reconstructed by the data analysis algorithm based on the 410 nm images captured at the same time as the white light images. The Gazelle-Multispectral data analysis algorithm sensitively and accurately identified Hb variants agreeing with HPLC reported results. Column 1–4: Multispectral test results for samples with different phenotypes. Column 1: Hb FA (Healthy newborn, Hb A: 8% vs. 6%, Hb F: 92% vs. 94%, Gazelle-Multispectral vs. HPLC); Column 2: Hb FS (Newborn with sickle cell disease, Hb F 83% vs. 89%, Hb S 17% vs. 11%, Gazelle-Multispectral vs. HPLC); Column 3: Hb FAS (Newborn with sickle cell trait, Hb S 16% vs. 16%, Hb F 57% vs. 55%, Hb A 27% vs. 29%, Gazelle-Multispectral vs. HPLC); and Column 4: Hb FAC (Newborn with Hb C disease, Hb C 20% vs. 20%, Hb F 45% vs. 45%, Hb A 35% vs. 35%, Gazelle-Multispectral vs. HPLC). Gazelle-Multispectral enabled identification and quantification of low concentration Hb variants with higher sensitivity (I–T) compared to white light imaging mode (A-H). †: Gazelle-Multispectral reports Hb C/E/A2 as demonstrated in Figure 1E. The identified Hb C/E/A2 were recognized as Hb C according to test location (Ghana).