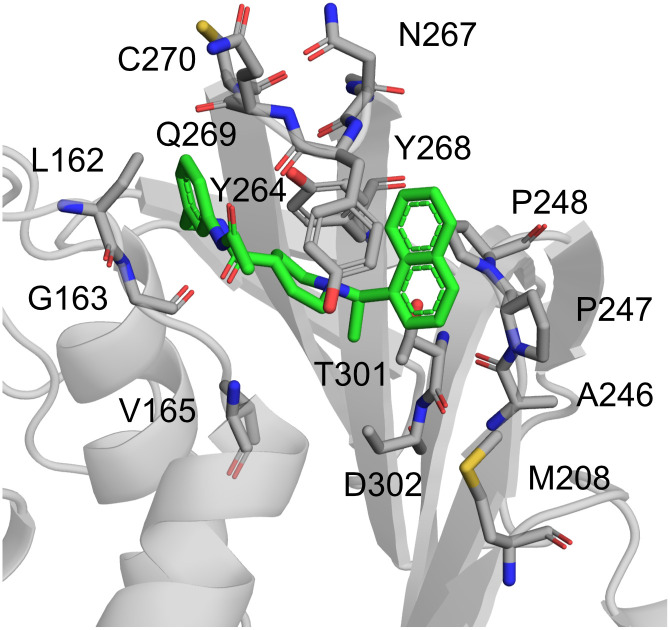
Fig 2. Structure of PLpro and variants within noncovalent inhibitor binding site.
The rates of mutations of these residues are presented in Table 1, along with the residues they are replaced to. It is worth noting that the residues forming the BL2 also can be mutated (aa 267–270). Based on PLpro-S43 complex from PDB ID: 7e35.