Abstract
Tumor suppressor mutations in head and neck squamous cell carcinoma (HNSCC) dominate the genomic landscape, hindering the development of effective targeted therapies. Truncating and missense mutations in NOTCH1 are frequent in HNSCC, and inhibition of PI3K can selectively target NOTCH1 mutant (NOTCH1MUT) HNSCC cells . In this study, we identify several proteins that are differentially regulated in HNSCC cells after PI3K inhibition based on NOTCH1 mutation status. Expression of Aurora kinase B (Aurora B), AKT, and PDK1 following PI3K inhibition was significantly lower in NOTCH1MUT cell lines than in NOTCH1WT cells or NOTCH1MUT cells with acquired resistance to PI3K inhibition. Combined inhibition of PI3K and Aurora B was synergistic, enhancing apoptosis in vitro and leading to durable tumor regression in vivo. Overexpression of Aurora B in NOTCH1MUT HNSCC cells led to resistance to PI3K inhibition, while Aurora B knockdown increased sensitivity of NOTCH1WT cells. Additionally, overexpression of Aurora B in NOTCH1MUT HNSCC cells increased total protein levels of AKT and PDK1. AKT depletion in NOTCH1WT cells and overexpression in NOTCH1MUT cells similarly altered sensitivity to PI3K inhibition, and manipulation of AKT levels affected PDK1 but not Aurora B levels. These data define a novel pathway in which Aurora B upregulates AKT that subsequently increases PDK1 selectively in NOTCH1MUT cells to mediate HNSCC survival in response to PI3K inhibition. These findings may lead to an effective therapeutic approach for HNSCC with NOTCH1 mutations while sparing normal cells.
Keywords: NOTCH1, HNSCC, Aurora kinase, PI3K, AKT
INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is common, lethal, and disfiguring cancer with no biomarker-selected, molecularly targeted therapies available. The most recent genomic information available for this disease has not been translated into clinical care largely because the genomic landscape is dominated by tumor suppressors, including NOTCH1, which is mutated in about 18% of HNSCCs (1-4). The pattern of truncating and missense NOTCH1 mutations and supporting laboratory data demonstrate its role as a tumor suppressor in HNSCC (5).
To target genomic alterations in HNSCC, we recently assessed the degree to which responses to PI3K/mTOR pathway inhibitors were associated with gene mutations, mRNA and protein expression in 59 HNSCC cell lines. Remarkably, HNSCC cell lines harboring NOTCH1 mutations (NOTCH1MUT) were significantly more sensitive to six drugs targeting PI3K or PI3K/mTOR than wild-type (WT) NOTCH1 cell lines (6). Additionally, NOTCH1MUT HNSCC cells treated with PI3K inhibitors underwent significant apoptosis in vitro and in vivo. In contrast, HNSCC cells with PIK3CA mutations exhibited growth arrest, but not cell death, when treated with PI3K inhibitors (7). A clinical trial testing a dual PI3K/mTOR inhibitor in patients with NOTCH1MUT HNSCC with recurrent or metastatic disease showed modest single-agent clinical activity (NCT03740100, (8))
Modest clinical responses and acquired resistance (AR) are leading causes of failure for otherwise promising and well-tolerated molecular targeted therapies, highlighting the importance of understanding molecular mechanisms of drug resistance. Furthermore, mechanisms of AR may overlap with innate resistance in patients with suboptimal initial responses. In this regard, potential mechanisms of resistance to PI3K inhibitors in NOTCH1WT HNSCC remain unknown, and this represents a major gap in knowledge. We hypothesized that differentially regulated pathways following PI3K inhibition in sensitive and resistant HNSCC cell lines mediate resistance. In the current study, we determined the expression of over 300 proteins and phosphoproteins in both sensitive NOTCH1MUT and resistant NOTCH1WT HNSCC cell lines after PI3K inhibition to identify the key downstream pathways involved in PI3K inhibitor-induced apoptosis. We then examined whether NOTCH1WT HNSCC could be sensitized to PI3K inhibition through simultaneously targeting of key downstream signaling pathways that mediate apoptosis in drug-sensitive NOTCH1MUT HNSCC. We further tested whether these combination therapies could maximize killing of drug-naïve NOTCH1MUT HNSCC and overcome AR that develops after prolonged single-agent PI3K inhibitor treatment.
Aurora kinase B (Aurora B, AURKB) was identified as a key effector molecule downstream of PI3K which was downregulated in drug-naïve NOTCH1MUT HNSCC compared with drug-resistant derivatives. Co-targeting Aurora B together with PI3K enhanced killing of drug-naïve tumors, including NOTCH1WT HNSCC, and could reverse AR in NOTCH1MUT HNSCC. Furthermore, we demonstrated a link between downregulation of Aurora B and PDK1, another important mediator of PI3K inhibition-induced apoptosis we previously identified (6).
To the best of our knowledge, this is the first study to identify Aurora B as a mechanism of resistance to PI3K inhibition in any cancer type. Because NOTCH1 loss-of-function mutations are common in other squamous cell carcinomas, including those of the skin (9), esophagus (10, 11), and lung (12), our findings may be broadly applicable to many patients.
MATERIALS AND METHODS
Cell culture
A panel of 56 HNSCC were obtained and maintained in their respective growth media as previously described (13-15). HNSCC cell lines used extensively in this study- HN31, UMSCC22A, PCI-15B, FaDu, MDA1386TU were a kind gift from Dr. Jeffrey Myers (MD Anderson Cancer Center) whereas HEK293 were purchased from ATCC. These cell lines were cultured in high glucose DMEM supplemented with 10% FBS, 1% Penicillin and Streptomycin and 2mM L-Glutamine and maintained in a humidified incubator with 5% CO2. All cell lines were genotyped by short tandem repeat analysis and were mycoplasma-free at the time of testing according to the Mycoplasma Detection Kit (LT07-705, Lonza, Walkersville, MD).
RPPA analysis
We measured protein levels using reverse phase protein array (RPPA) with a panel of 304 antibodies (Supplementary Table S1) (16) as previously described and performed immunoblotting as explained briefly below. For RPPA data processing, scanned images after hybridization and labeling were quantified using commercial software, MicroVigene (http://www.vigenetech.com/MicroVigene.htm) and SuperCurve (17). Our method iteratively fit joint logistic models to all data on a slide and returned summary estimates of log2 protein expression values for each sample. The expression values were normalized across slides using a median centering approach to adjust for variability in sample loading, inducing consistent differences affecting all arrays in a set.
Plasmids and reagents
Doxycycline-inducible lentiviral vector for FOXM1b expression in mammalian cells (pCW57.1-FOXM1b) was obtained from Addgene. HA-tagged Aurora B plasmid was a kind gift from Dr. Ramon Parsons (Icahn School of Medicine, Mt. Sinai, NY). The drugs used in the current study were purchased form Selleck Chemicals (Houston, TX) and MedChem Express (NY; Supplementary Table S2).
Animal studies
All animal studies were approved by the Institutional Animal Care and Use Committee at The University of Texas MD Anderson Cancer Center and are detailed in the supplementary methods. PASS 13 power analysis and sample size software (2014; NCSS, LLC, Kaysville, UT) was used for the power/sample-size analysis, and investigators carried out these studies unblinded. Subcutaneous xenograft models were generated by injecting NOTCH1MUT cells into athymic nude mice. Briefly, HN31 (0.75 × 106 cells) or UMSCC22A (3 × 106 cells) were injected subcutaneously on the right flank of the mice. Once the tumor volume reached ≥60 mm3, mice were randomized in their respective treatment groups. Mice injected with UMSCC22A cells were treated intraperitoneally with 10 mg/kg copanlisib (BAY806946) three times per week. Mice bearing HN31 tumors received 14 mg/kg copanlisib (BAY841236) intraperitoneally, 30 mg/kg alisertib by oral gavage, 50mg/kg barasertib intraperitoneally, or a combination of copanlisib with alisertib or barasertib for 5 days per week for 21 days.
Apoptosis assay
Apoptosis assays were performed as described previously (6). Briefly, TUNEL staining was carried out with either APO-BRDU (556405, BD Biosciences, San Diego, CA) or APO-DIRECT kit (556381, BD Biosciences), and Annexin V/PI staining was performed with a FITC-Annexin V apoptosis detection kit (556547, BD Biosciences) or PE-Annexin V apoptosis kit (559763, BD Biosciences) at indicated times. Samples were processed at the MD Anderson Flow Cytometry and Cell Imaging Core laboratory with a 3-laser, 10-color Gallios flow cytometer (Beckman Coulter) and analyzed using Kaluza software (Beckman Coulter).
Western Blotting
Cells were lysed with ice-cold 1X RIPA buffer supplemented with protease and phosphatase inhibitors, and the lysates were centrifuged at 20,000g for 10 minutes at 4 °C. Lysates containing equal amounts of protein were resolved using SDS-PAGE, transferred to PVDF membranes, and incubated with primary antibody overnight. Protein expression was detected using a horseradish peroxidase–conjugated secondary antibody (Bio-Rad) and electrochemiluminescence reagent (32106, ThermoFisher Scientific; or 1705062, Bio-Rad). The primary antibodies used in the study are listed in Supplementary Table S3.
RNA sequencing
RNA was isolated using the Qiagen RNeasy kit after treating cells with 200nM copanlisib or vehicle for 18 hours. Biological triplicate RNA samples underwent next-generation sequencing at Psomagen to generate an average of approximately 9 million reads per sample, which were mapped, aligned, and quantitated using the RSEM method. Upper quartile normalized fragments per kilobase of transcript per million mapped reads (FPKMUQ) values were then calculated using the formula
Log2 FPKMUQ values were calculated, and replicate samples were grouped by cell line and treatment. Low-expression genes were removed by filtering out any gene whose maximum group average across all treatments and cell lines was <2 (log2 space). Differences in gene expression between drug-treated and vehicle-treated values for each cell line were examined for statistical significance by performing multiple t tests and applying the Benjamini-Hochberg correction to control the false discovery rate (FDR), and differences with an FDR <0.1 were considered significant. Genes with significant differences were dichotomized into those upregulated (i.e., increased) or downregulated (i.e., decreased) following treatment with copanlisib to identify disjoint and common differentially expressed genes across cell lines.
Cell viability assay
As described previously (6, 16), cells were treated with DMSO or inhibitors as indicated at seven different concentrations (0-2μM) for 72 hours. Luminescence was measured using CellTiter-Glo (G7570, Promega, Madison, WI) according to the manufacturer’s instructions. For the combination screening in 56 HNSCC cell lines, the drexplorer R package with a best fit dose response model was used to calculate inhibitory concentration values (18). The CI values were calculated using the Chou-Talalay method (19-21) using Calcusyn software (Biosoft). CI values <1 are considered synergistic.
Colony formation assay
Colony formation assay was performed in triplicates as previously described (6). Briefly, cells were seeded in 6-well plates and treated with DMSO or the indicated drugs. After 48 hours of treatment, media was replaced with drug-free media and cells were cultured for 14-21 days. The cell colonies were fixed and stained with crystal violet and analyzed using image J software (NIH, Bethesda, MD).
Reactive oxygen species experiments
CellROX™ Green Flow Cytometry Assay kit (C10492, ThermoFisher Scientific) was used for the detection of reactive oxygen species in live cells according to the manufacturer’s instructions.
Gene knockdown and overexpression
Cells expressing inducible FOXM1b were generated using lentiviral transduction and selected with 0.5μg/ml puromycin. FOXM1b expression was induced with 100ng/ml doxycycline. Cells constitutively expressing HA-tagged Aurora B, Aurora A, and AKT1 were generated using lentiviral particles obtained from the Functional Genomics Core laboratory at MD Anderson. Cells overexpressing HA-tagged Aurora B were selected with 2 μg/mL puromycin and cells with Aurora A and AKT1 overexpression were selected with 10 μg/mL blasticidin. For gene knockdown in HNSCC cells, siRNA was transfected with Lipofectamine RNAiMAX (13778-075, ThermoFisher Scientific) according to the manufacturer’s instructions. Two specific siRNAs directed against AURKB and AKT1 were obtained from Horizon Discovery (AurB (1) -L-003326-00-0005; AKT1 (1) -L-003000-00-0005, siRNA SMARTpools), Invitrogen (AurB (2) – 4390824) and Santa Cruz Biotechnology (AKT1 (2) – sc-44198).
Statistical analysis
RPPA data analyses were performed using R (version 2.6.3). The two experimental factors in the RPPA data were phenotypes (NOTCH1WT/NOTCH1MUT cell lines) and PI3K treatment (vehicle/omipalisib; Fig. 1A). The limma package in Bioconductor (https://bioconductor.org/packages/release/bioc/html/limma.html) was used to compare changes in NOTCH1WT and NOTCH1MUT cell lines after treatment, and changes between the two (to identify differentially expressed proteins for treatment between phenotypes), which was the interaction term of the linear model. The Benjamini-Hochberg method (22) was applied to the resulting P values to control FDR. All statistical analyses were performed using R version 4.0.1 (R Core Team, 2020) or GraphPad Prism 7. An unpaired t test was used to compare the mean of two different groups when the distribution of the population was normal. One-way and two-way analysis of variance were used to compare the means of three or more groups under the assumption of normal distribution, and the Dunnett test or Tukey honestly significant difference test was applied for multiple comparisons. Kruskal-Wallis rank sum test was used to compare the means of more than two groups which didn’t follow normal distribution. The Wilcoxon rank sum tests with Benjamini & Hochberg method were used to compare the pairwise groups after Kruskal-Wallis. All P values were two-tailed and for all analyses, P ≤ 0.05 was considered statistically significant, unless otherwise specified.
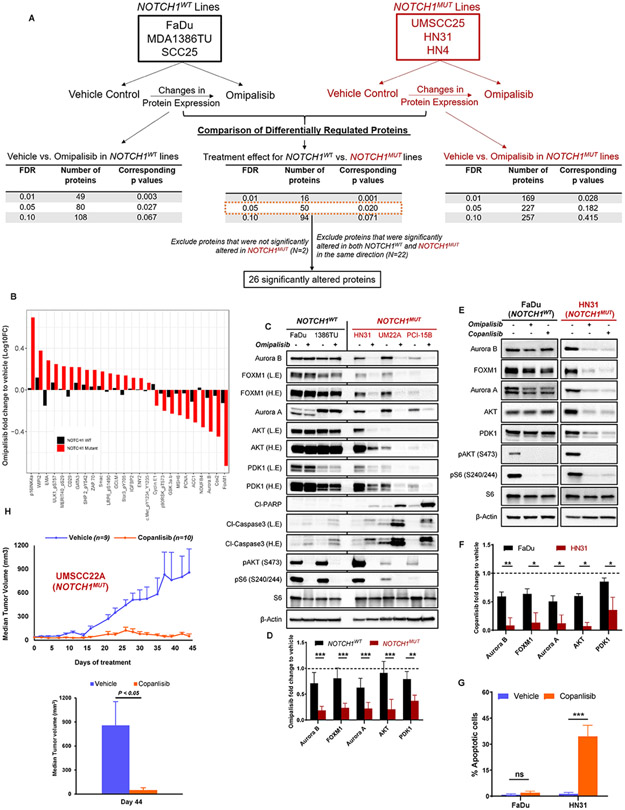
Fig. 1. Aurora kinases, AKT, PDK1, and FOXM1 are downregulated in NOTCH1 mutant (NOTCH1MUT) head and neck squamous cell carcinoma (HNSCC) cells upon PI3K inhibition.
(A) Schematic showing experimental design and the number of differentially expressed proteins according to reverse phase protein array (RPPA) analysis in NOTCH1WT and NOTCH1MUT HNSCC upon PI3K/mTOR inhibition using 50nM omipalisib at 24 hours. FDR, false discovery rate. (B) Waterfall plot showing the 26 differentially expressed proteins that were significantly altered as a treatment effect of omipalisib as determined by RPPA. Immunoblots of differentially expressed proteins and apoptosis markers in NOTCH1WT and NOTCH1MUT HNSCC cell lines when treated for 24 hours with 50nM omipalisib (C) or 200nM copanlisib (E). (D,F) Protein quantification of (C-E) by Image J, normalized using β-Actin as a control, and subsequent fold change to treatment with vehicle. Cl, cleaved; L.E, low exposure; H.E, high exposure. Bars indicate mean ± SD from two NOTCH1WT cell lines and three NOTCH1MUT cell lines and three biological replicates: *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed Student t test. (G) Apoptosis measured by FITC-dUTP/propidium iodide staining in FaDu and HN31 cells treated with 200nM copanlisib for 48 hours. The percentages of apoptotic cells are expressed as the mean ± SD from three independent experiments: ***P < 0.001, ns, nonsignificant, two-tailed Student t test. (H) The NOTCH1MUT cell line UMSCC22A was injected subcutaneously into athymic nude mice. After tumors reached >60 mm3, the mice were randomized to receive either vehicle control (PEG400/acidified water) or 10 mg/kg copanlisib (BAY806946) by intraperitoneal injection three times per week for 6 weeks. Tumor sizes were measured, and tumor volumes were calculated at the indicated times and plotted ±SEM (unpaired Student t test), n, number of mice examined in each group.
Data availability
The data generated in this study are available within the article and its supplementary data files. Derived data supporting the findings of this study are available from the corresponding author upon request.
RESULTS
PI3K inhibition leads to differential expression of Aurora kinases and AKT in NOTCH1MUT and NOTCH1WT HNSCC cell lines
To investigate potential mechanisms mediating resistance to PI3K inhibition, we measured the levels of 304 proteins and phosphoproteins using RPPA analysis in three resistant NOTCH1WT (FaDu, MDA1386TU, SCC25) and three sensitive NOTCH1MUT (HN31, HN4, UMSCC25) cell lines after 4 and 24 hours of treatment with 50nM omipalisib (Fig. 1A), which is below its peak plasma concentration of 90nM (23). Omipalisib is a potent dual PI3K/mTOR inhibitor that was in clinical development at the time RPPA was performed. Many proteins were affected by PI3K/mTOR inhibition in both NOTCH1MUT and NOTCH1WT cell lines, including the expected changes in the PI3K/AKT/mTOR pathway, confirming appropriate and durable pathway inhibition (Supplementary Fig. S1A).
After 24 hours of treatment, 16 proteins were differentially expressed as a result of omipalisib treatment and NOTCH1 mutation status (interaction P < 0.05 for treatment effect for NOTCH1WT and NOTCH1MUT) at FDR of 0.01, and 50 proteins were differentially expressed at FDR of 0.05 (Supplementary Fig. S1B, S2A). Differentially regulated proteins included expected markers of apoptosis (Mcl1, MDM2) and proliferation (p16, cyclin E1, PCNA, CDK1, Rb) in addition to Aurora B, forkhead box M1 (FOXM1), and several proteins involved in metabolism (glutamate D1, G6PD, ACC1). After 4 hours of treatment, 10 proteins were differentially regulated at an FDR of 0.05 (Supplementary Fig. S1C), but we did not study these because the magnitude of changes in these proteins was small and similar in NOTCH1WT and NOTCH1MUT cells.
For further study, we focused on proteins that were significantly altered after omipalisib treatment only in the NOTCH1MUT cell lines and proteins that were regulated in opposite directions in the NOTCH1WT and NOTCH1MUT cell lines (Fig. 1B). We used immunoblotting to validate the RPPA findings in two NOTCH1WT (FaDu, MDA1386TU) and three NOTCH1MUT (HN31, UMSCC22A, PCI-15B) cell lines and confirmed consistent differences in the regulation of Aurora B and FOXM1 based on NOTCH1 mutational status following treatment with 50nM omipalisib for 24 hours (Fig. 1C, D). We also examined Aurora kinase A (Aurora A, AURKA), although it was not included in the RPPA, because Aurora A is known to regulate Aurora B (24). We detected similar changes in Aurora A protein levels in all NOTCH1MUT HNSCC cell lines (Fig. 1C, D).
A recent study demonstrating destabilization of Aurora B levels through AKT degradation in PI3K/PTEN pathway mutant and WT KRAS/BRAF cancers (25) prompted us to investigate the total levels of AKT as well. We identified a remarkable decrease in total AKT levels across all NOTCH1MUT HNSCC cell lines upon PI3K/mTOR inhibition (Fig. 1C, D). Consistent with our previous findings, we found both a significant downregulation in PDK1 levels (Fig. 1C, D) and evidence of apoptosis in NOTCH1MUT lines treated with omipalisib (Fig. 1C) (6). Moreover, RPPA showed significant changes in the levels of AKT and phospho-PDK1 selectively in NOTCH1MUT HNSCC cell lines, although the interaction P value was >0.05 for total AKT (Supplementary Fig. S2B). In contrast, several proteins identified by RPPA were differentially regulated in some, but not all, of the cell lines based on NOTCH1 mutation status according to immunoblotting (Supplementary Fig. S1D).
To determine if these expression changes are specific to PI3K and confirm the robustness of our findings, we treated NOTCH1WT and NOTCH1MUT HNSCC cell lines in parallel with omipalisib or copanlisib, which is a potent, well-tolerated (26) pan-PI3K and US Food and Drug Administration–approved drug. Both omipalisib and copanlisib reduced levels of Aurora A/B, FOXM1, PDK1, and total AKT to a much greater extent in NOTCH1MUT than in NOTCH1WT cell lines (Fig. 1E, F). Additionally, copanlisib caused a significant increase in apoptosis in NOTCH1MUT cells (Fig. 1G). Furthermore, we assessed the antitumor efficacy of copanlisib in a subcutaneous model of NOTCH1MUT HNSCC (UMSCC22A) and found significantly lower tumor volumes compared with the vehicle-treated mice (Fig. 1H). These findings confirm that both pan-PI3K and dual PI3K/mTOR inhibitors cause apoptosis in NOTCH1MUT HNSCC cells in vitro (6) and show that pan-PI3K inhibition is effective in vivo. For subsequent mechanistic experiments, we chose to continue with the more specific and clinically relevant drug, copanlisib.
Pan-PI3K inhibition leads to reduced Aurora kinase and AKT levels selectively in NOTCH1MUT HNSCC
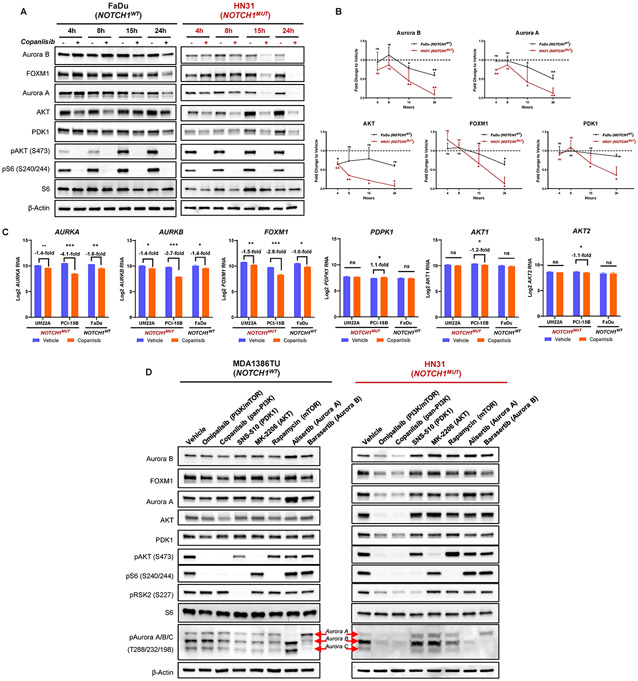
We determined the dynamics of the differentially altered proteins through time-course analyses. Although the PI3K pathway was inhibited as early as 4 hours after drug treatment in all cell lines, protein levels of Aurora A/B, total AKT, FOXM1, and PDK1 were not significantly decreased until 15 hours (Fig. 2A, B; Supplementary Fig. S3A-C), and further decreased through 24 hours of drug treatment in the NOTCH1MUT lines. Furthermore, PI3K inhibition led to a more profound and sustained effect on the levels of these proteins in the NOTCH1MUT lines than in the NOTCH1WT lines (Fig. 2B; Supplementary Fig. S3B, C). In contrast, although AKT protein levels initially dropped modestly in NOTCH1WT cells at 4-8 hours, AKT levels stabilized at 15 and 24 hours after PI3K inhibition in these cells (Fig. 2A, B; Supplementary Fig. S3A-C). Altogether, these data show that apoptosis in NOTCH1MUT HNSCC cell lines correlates with decreased levels of Aurora A/B, FOXM1, AKT, and/or PDK1 as a result of PI3K pathway inhibition.
Fig. 2. PI3K inhibition results in considerable proteomic alterations selectively in NOTCH1 mutant (NOTCH1MUT) head and neck squamous cell carcinoma (HNSCC).
(A) Representative Western blot analysis images of NOTCH1WT and NOTCH1MUT HNSCC cell lines at indicated time points after treatment with 200nM copanlisib (h, hours). (B) Quantification of the Western blot images shown in (A) from at least two independent experiments (mean ± SD); ns, nonsignificant, *P < 0.05, **P < 0.01, two-sample t test. (C) Graphs showing mRNA levels measured by RNA sequencing of indicated genes in NOTCH1MUT (UMSCC22A, PCI-15B) and NOTCH1WT (FaDu) cell lines after treatment with 200nM copanlisib at 18 hours. Data are presented as the mean ± SD of at least three independent experiments; *P < 0.05, **P < 0.01, ***P < 0.001, ns, nonsignificant, multiple t-tests with Benjamini-Hochberg correction. (D) Western blot analysis of protein changes in NOTCH1WT and NOTCH1MUT HNSCC cell lines after treatment with indicated drugs (50nM omipalisib, 50nM barasertib, 200nM copanlisib, 200nM alisertib, 250nM rapamycin, 300nM SNS-510, 400nM MK-2206) for 24 hours.
We next determined if these proteomic alterations were at the transcriptional level using RNA sequencing in two NOTCH1MUT and one NOTCH1WT cell line. We found a significant reduction in AURKA, AURKB, and FOXM1 mRNA levels in all HNSCC cells treated with copanlisib for 18 hours (Fig. 2C). In contrast, PDPK1 (PDK1), AKT1 and AKT2 mRNA levels were unaffected by PI3K inhibition, suggesting that the changes in their protein levels are post-translational (Fig. 2C).
To gain further insight into how these concordantly regulated proteins interact with the PI3K pathway, we inhibited the individual molecules using kinase inhibitors: SNS-510 (PDK1), MK-2206 (AKT), rapamycin (mTOR), alisertib (Aurora A), and barasertib (Aurora B) at target-specific concentrations. As expected, protein levels of PDK1, AKT, FOXM1, and Aurora A/B decreased substantially following treatment with the dual PI3K/mTOR inhibitor (omipalisib) and pan-PI3K inhibitor (copanlisib) only in NOTCH1MUT cells (Fig. 2D; Supplementary Fig. S3D-E). However, other kinase inhibitors targeting the PI3K pathway, including those affecting PDK1, mTOR, AKT, or PI3Kα (Supplementary Fig. S3E), did not affect levels of Aurora A/B, FOXM1, AKT, or PDK1. Likewise, Aurora A/B kinase inhibition also did not affect levels of any of these proteins (Fig. 2D; Supplementary Fig. S3D, E).
Concurrent inhibition of Aurora A/B and PI3K is synergistic in HNSCC cell lines in vitro and in vivo
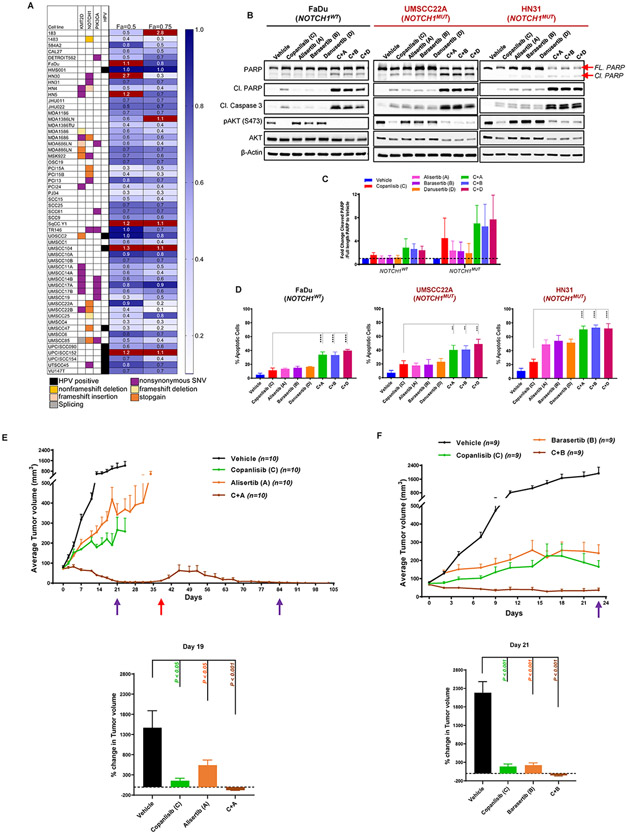
Given its differential regulation, we hypothesized that the maintenance of Aurora A/B expression in NOTCH1WT HNSCC contributed to resistance to PI3K inhibition. To test this hypothesis, we combined the pan-Aurora inhibitor danusertib (0-2μM) with the dual PI3K/mTOR inhibitor omipalisib (0-200nM) at a fixed 1:1 ratio in 56 HNSCC cell lines for 72 hours and tested cell viability. The combination index (CI) was less than 1, indicating synergy, in 46 of 56 HNSCC cell lines (82%) at an effect size of 0.5 and in 49 of 56 cell lines (88%) at an effect size of 0.75 (Fig. 3A). Among the 13 NOTCH1MUT HNSCC cell lines, all had CI values less than 1 at an effect size of 0.75, suggesting that inhibiting the residual Aurora kinases in NOTCH1MUT can also enhance cell death.
Fig. 3. Combined targeting of PI3K and Aurora A/B enhances apoptosis and decreases tumor size of head and neck squamous cell carcinoma (HNSCC).
(A) Heat map depicting the combination index values in 56 HNSCC cell lines upon combined treatment with danusertib (0-2μM) and omipalisib (0-200nM) at a fixed 1:1 ratio at effect sizes (Fa) 0.5 and 0.75. (B) Representative Western blot analysis from three independent experiments in NOTCH1WT (FaDu) and NOTCH1MUT (UMSCC22A, HN31) HNSCC cell lines treated with indicated drugs (200nM copanlisib, 200nM alisertib, 50nM barasertib, 500nM danusertib) either as single agent or in combination for 24 hours. Cl, cleaved, FL, full length. (C) Ratio of cleaved PARP to full-length PARP protein expression from the Western blot analysis shown in (B), quantified using ImageJ and normalized to β-Actin as a loading control, and the relative fold control from vehicle. Bars indicate mean ± SD from three independent experiments in NOTCH1WT (FaDu) cell line and NOTCH1MUT (UMSCC22A, HN31) cell lines. Dashed line indicates 1-fold change in the ratio of cleaved PARP to full-length PARP. (D) Percentage of apoptotic cells measured from Annexin V/propidium iodide staining in FaDu, UMSCC22A, and HN31 cells upon treatment with indicated drugs (200nM copanlisib, 200nM alisertib, 50nM barasertib, 500nM danusertib) either as single agent or in combination for 24-36 hours. The percentage of apoptotic cells is expressed as the mean ± SD from three independent experiments; **P < 0.01, ***P < 0.001, ****P < 0.0001, one-way analysis of variance corrected for multiple comparisons with the Tukey test. (E, F) The NOTCH1MUT cell line HN31 was injected subcutaneously into athymic nude mice. After tumors reached 60 mm3 , the mice were randomized to receive either vehicle (0.9% NaCl, 2-hydroxypropyl-β-cyclodextrin or 1M Tris pH 9.0), 14 mg/kg copanlisib (BAY 841236) by IP, 30mg/kg alisertib (MLN8237) by oral gavage (E) or 50mg/kg barasertib (AZD1152) by IP (F) and a combination of copanlisib with alisertib (E) or barasertib (F) for five days a week for 3 weeks. Tumor sizes were measured, and tumor volumes plotted ± SEM and the indicates times, P-values were calculated using one-way ANOVA Kruskal-Wallis rank sum test followed by pairwise Wilcoxon rank sum tests, Type I error rate is controlled by Benjamini & Hochberg method (FDR). Purple arrows indicate end of treatment and red arrow indicates restart of drug treatment, n-number of mice examined in each group.
We also tested the effects of more specific inhibitors of PI3K (copanlisib), Aurora A (alisertib), and Aurora B (barasertib) at clinically relevant concentrations. Alisertib (MLN8237) inhibits catalytic activity of Aurora A, and at higher concentrations can also inhibit Aurora B both in vitro and in vivo (27). Barasertib (AZD2811, AZD1152) is a potent and selective inhibitor of Aurora B (28, 29) that is currently in clinical development (NCT02579226). We treated NOTCH1WT and NOTCH1MUT HNSCC cell lines with alisertib, barasertib, or danusertib alone or combined with copanlisib for 24 hours and detected significantly increased induction of apoptosis as measured by cleaved PARP, cleaved caspase 3, and Annexin V and propidium iodide (PI) staining in the combinations compared with the single agents (Fig. 3B-D Supplementary Fig. S4A-D). We found varying sensitivities to Aurora A/B inhibitors alone across all cell lines, with HN31 exhibiting the highest sensitivity and FaDu the lowest. However, the combined inhibition of PI3K and Aurora A/B not only led to increased apoptosis of NOTCH1MUT HNSCC cell lines but also sensitized otherwise resistant NOTCH1WT HNSCC cell lines. Furthermore, when we combined barasertib with omipalisib or copanlisib in four NOTCH1WT cell lines for 72 hours and measured cell viability, the CI values were less than 1 in all four cell lines, indicating synergy (Supplementary Fig. S4E). We used the HEK293 cell line as a non-transformed control and found no significant apoptosis with either single agents or combined PI3K and Aurora kinase inhibition (Supplementary Fig. S4 F, G). These in vitro findings strongly suggest that combined inhibition of PI3K and Aurora A/B enhanced PI3K-induced apoptosis of NOTCH1MUT HNSCC cell lines and sensitized NOTCH1WT HNSCC cell lines.
We then tested these combinations in vivo using a xenograft model of NOTCH1MUT HNSCC (HN31) and administered copanlisib and alisertib or barasertib for 21 days. When compared with vehicle-treated group (1466% ± 422%; Fig. 3E, Supplementary Fig. S4H), mice receiving copanlisib or alisertib alone demonstrated significantly smaller tumor volumes (copanlisib: 156% ± 61%, P < 0.05; alisertib: 540% ± 129%, P < 0.05) compared to baseline, whereas the combined treatment led to tumor regression (−78% ± 6%, P < 0.01) at day 19. Similarly, mice treated with a combination of copanlisib and barasertib (−51% ± 18%, P<0.001; Fig 3F, Supplementary Fig. S4I) showed significant reduction in tumor size, whereas mice receiving copanlisib or barasertib alone exhibited substantial smaller tumor volumes (copanlisib: 187% ± 69%, P<0.001; barasertib: 223% ± 61%, P<0.001) when compared to the vehicle treated group (2182% ± 304%) at day 21. However, alisertib was better tolerated than barasertib and therefore the mice treated with the combination of alisertib and copanlisib underwent a second cycle of treatment and exhibited prolonged and durable tumor regression (Fig. 3E, Supplementary Fig. S4H).
Aurora kinases mediate AR to PI3K inhibition
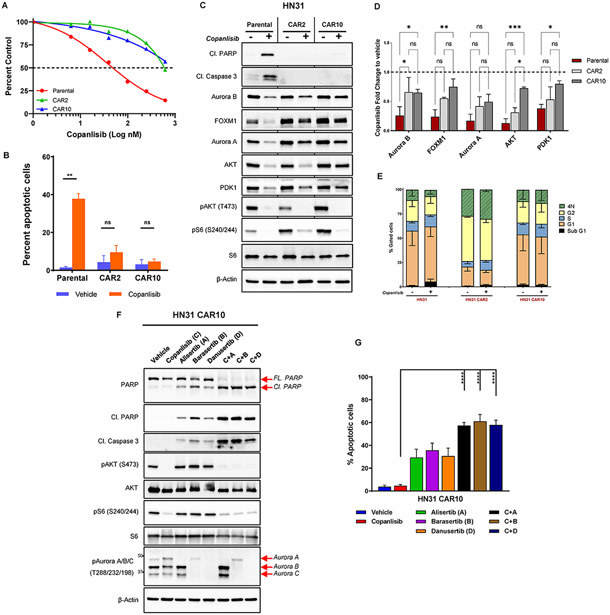
To investigate mechanisms of AR, we exposed the HN31 cell line to increasing concentrations of copanlisib over time until resistance emerged. Subsequently, single-cell clones were established after cell sorting, and the resulting clones (CAR2, CAR10) were tested for sensitivity to copanlisib and omipalisib by cell viability assay and FITC-dUTP/PI staining (Fig. 4A, B; Supplementary Fig. S5A, B). The AR clones had a significant shift in half-maximal inhibitory concentrations (IC50) compared with the parental cells (Fig. 4A, Supplementary Fig. S5A) and did not exhibit any significant apoptosis or changes in the cell cycle upon PI3K inhibition (Fig. 4B, C, E; Supplementary Fig. S5B). Moreover, following PI3K/mTOR inhibition, total levels of Aurora B, PDK1, AKT, and FOXM1 decreased more substantially in the parental cells than in the AR clones (Fig. 4C, D; Supplementary Fig. S5C). However, the AR clones still retained copanlisib-induced changes in these proteins, suggesting that they may also engage in additional, novel mechanisms of resistance. To determine whether these protein changes were as a result of changes at the mRNA level, we conducted RNA sequencing in the AR clones and HN31 parental cells. Similar to the findings from other NOTCH1MUT cell lines UMSCC22A and PCI-15B in Fig. 2C, we found > 2 fold reduction in AURKA, AURKB, and FOXM1 mRNA levels in the parental cells but not in the AR clones treated with copanlisib for 18 hours (Supplementary Fig. S5D). Furthermore, the mRNA levels of PDPK1 (PDK1), AKT1 and AKT2 mRNA levels were unaffected by PI3K inhibition in both parental and the AR clones, suggesting that the observed changes in their protein levels are post-translational (Supplementary Fig. S5D).
Fig. 4. Aurora kinases mediate acquired resistance to PI3K inhibition in NOTCH1 mutant (NOTCH1MUT) head and neck squamous cell carcinoma.
(A) HN31 parental and copanlisib acquired resistant (CAR) cells were treated with increasing concentrations of copanlisib for 72 hours, and cell viability was measured with the CellTiter-Glo assay. CAR2 and CAR10 represent two different clones of resistant cells, and the dotted line denotes 50% cell population. (B) Apoptotic cells measured by FITC-dUTP/propidium iodide (PI) staining in HN31 parental and CAR cells treated with 200nM copanlisib for 48 hours. The values indicate mean ± SD from three independent experiments: **P < 0.01, ns, nonsignificant, two-tailed Student t test. (C) Western blot analysis of HN31 parental and CAR cells upon treatment with 200nM copanlisib for 24 hours. (D) Quantification of protein expression from the Western blot analysis shown in (C) using Image J and normalized using β-Actin as a control; subsequent fold change to vehicle was calculated. Bars indicate mean ± SD of two biological replicates; *P < 0.05, **P < 0.01, two-way analysis of variance corrected for multiple comparisons with the Tukey test. (E) Cell cycle analysis of HN31 parental and CAR cells treated with 200nM copanlisib for 48 hours measured from PI staining as mean ± SD of three independent experiments. (F) Western blot analysis of HN31 CAR10 cells treated with indicated drugs (200nM copanlisib, 200nM alisertib, 50nM barasertib, 500nM danusertib) either as single agent or in combination for 24 hours. FL, full length; Cl, cleaved. (G) Percentage of apoptotic cells measured from Annexin V/PI staining in HN31 CAR10 cells upon treatment with indicated drugs (200nM copanlisib, 200nM alisertib, 50nM barasertib, 500nM danusertib) for 24 hours. The percentage of apoptotic cells is expressed as the mean ± SD from three independent experiments; ***P < 0.001, ****P < 0.0001, one-way analysis of variance corrected for multiple comparisons with the Tukey test.
To test the hypothesis that mechanisms of innate resistance in NOTCH1WT HNSCC and AR in NOTCH1MUT HNSCC may both depend upon Aurora kinases, we inhibited PI3K and Aurora kinases simultaneously in the AR clones and analyzed for apoptosis. We observed increased cleavage of PARP and caspase 3 and higher Annexin V staining (Fig. 4F, G; Supplementary Fig. S5E, F) in the cells treated with combined inhibitors compared with vehicle or single-agent copanlisib. Sensitivity to single-agent Aurora kinase inhibitors appeared to be reduced in the AR clones compared with the parental cells, indicating that resistance to PI3K inhibition may alter sensitivity to Aurora kinase inhibitors (i.e., comparing parental HN31 from Fig. 3D with CAR data from Fig. 4G and Supplementary Fig. S5F).
FOXM1, reactive oxygen species, and Aurora A do not mediate PI3K inhibition-induced apoptosis in HNSCC
AKT positively regulates the oncogenic transcription factor FOXM1 and phosphorylates a FOXM1 inhibitor (FOXO3a) on a negative regulatory site that was affected after PI3K inhibition only in NOTCH1MUT cells (Supplementary Fig. S6A). These data led us to hypothesize that the canonical regulation of FOXO3a and FOXM1 is uncoupled from AKT activation in NOTCH1WT cells, explaining their drug resistance. In support of this hypothesis, FOXM1 can positively regulate genes required for mitosis (30), glycolysis (31, 32), and reactive oxygen species homeostasis (33) so disruption to any or all these processes may contribute to loss of viability. Consistent with this model, significant differences between drug-sensitive NOTCH1MUT and resistant NOTCH1WT cell lines were observed in drug-induced levels of key enzymes regulating glucose metabolism and cellular redox homeostasis (Supplementary Fig. S1B). To test this hypothesis, we manipulated levels of FOXM1 and scavenged reactive oxygen species in HNSCC cells treated with omipalisib and determined the effects on cell survival. Knockdown of FOXM1 in NOTCH1WT cells did not sensitize them to omipalisib-mediated apoptosis (Supplementary Fig. S6B). Overexpression of FOXM1 did not rescue NOTCH1MUT cells from omipalisib-induced apoptosis (Supplementary Fig. S6C, D). Although reactive oxygen species increased after treatment with omipalisib in NOTCH1MUT cells and was effectively scavenged by N-acetyl cysteine, treatment with N-acetyl cysteine did not rescue apoptosis in NOTCH1MUT cells (Supplementary Fig. S6E-G).
It was previously reported that Aurora A contributes to resistance to PI3K inhibition in breast cancer (34). We examined this possibility in our HNSCC models by overexpressing Aurora A in NOTCH1MUT cells. Subsequent treatment with copanlisib for 24 hours did not reverse the apoptotic phenotype of NOTCH1MUT cells, suggesting that an alternate pathway is responsible for drug sensitivity (Supplementary Fig. S6H).
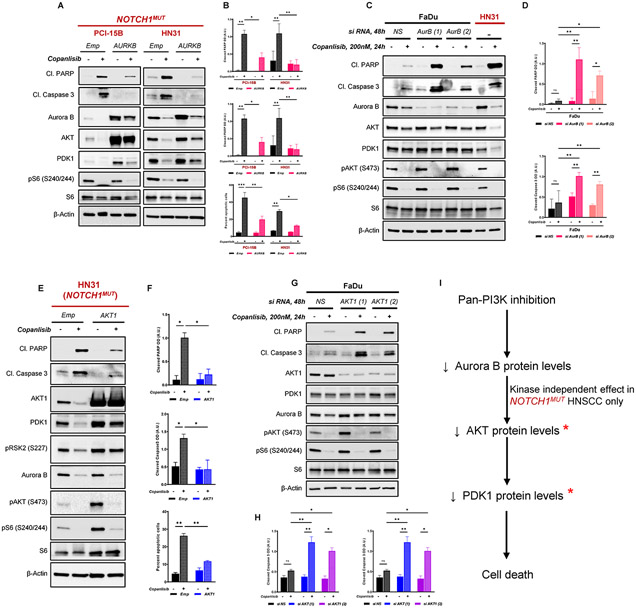
Aurora B dictates sensitivity to PI3K inhibition via regulation of AKT and PDK1 in NOTCH1MUT HNSCC
Because Aurora A and FOXM1 did not modulate sensitivity to PI3K inhibition in NOTCH1MUT HNSCC, we then tested the effect of altering Aurora B. Overexpression of Aurora B in NOTCH1MUT HNSCC cells partially rescued copanlisib-induced apoptosis, as demonstrated by markedly reduced cleaved PARP and caspase 3 proteins and Annexin V positive cells (Fig. 5A,B). AKT and PDK1 levels were also upregulated when Aurora B was overexpressed in NOTCH1MUT HNSCC (Fig. 5A). In NOTCH1WT HNSCC cells, siRNA-mediated Aurora B knockdown significantly enhanced PI3K inhibition-induced apoptosis (Fig. 5C,D, Supplementary Fig. S7A,B,C), but not to the same extent as we observed in NOTCH1MUT HNSCC (HN31) cells despite a similar level of Aurora B expression following treatment with copanlisib (Fig. 5C). Likewise, total levels of AKT and PDK1 were not significantly affected by Aurora B knockdown in NOTCH1WT HNSCC cells (Fig. 5C, Supplementary Fig. S7A).
Fig. 5. Aurora kinase B (Aurora B) protein levels dictate sensitivity to PI3K inhibition by stabilizing AKT and PDK1 levels.
(A) Representative Western blot analysis from three independent experiments of NOTCH1MUT (PCI-15B and HN31) HNSCC cells stably overexpressing empty vector (Emp) or Aurora B (AURKB) treated with 200nM copanlisib for 24 hours. (B) Bar graphs showing quantification of cleaved PARP, cleaved Caspase 3 from western blots in (A) and percent apoptotic cells as measured by Annexin V/PI staining in Aurora B overexpressing cells. *P < 0.05, **P < 0.01, ***P < 0.001, one-way analysis of variance corrected for multiple comparisons with the Tukey test. (C) Representative immunoblotting analysis from three independent experiments in NOTCH1WT (FaDu) cells depleted of Aurora B by siRNA-mediated knockdown using two specific siRNAs – AurB(1) and AurB (2) treated with 200nM copanlisib at indicated times. (D) Quantification of cleaved PARP, cleaved Caspase 3 from Western blots in (C). *P < 0.05, **P < 0.01, ns, nonsignificant; one-way analysis of variance corrected for multiple comparisons with the Tukey test. (E) Representative Western blot analysis from three independent experiments of NOTCH1MUT (HN31) HNSCC cells stably overexpressing empty vector (Emp) or AKT1 (AKT1) treated with 200nM copanlisib for 24 hours. (F) Quantification of cleaved PARP, cleaved Caspase 3 from Western blots in (E) and percent apoptotic cells as measured by Annexin V/7AAD staining in AKT1 overexpressing cells. *P < 0.05, **P < 0.01, one-way analysis of variance corrected for multiple comparisons with the Tukey test. (G) Representative immunoblotting analysis from three independent experiments in NOTCH1WT (FaDu) cells depleted of AKT1 by siRNA-mediated knockdown using two specific siRNAs – AKT1(1) and AKT1 (2) treated with 200nM copanlisib at indicated times. (H) Quantification of cleaved PARP, cleaved Caspase 3 from Western blots in (G). *P < 0.05, **P < 0.01, ns, nonsignificant; one-way analysis of variance corrected for multiple comparisons with the Tukey test. (I) Proposed model illustrating the mechanism of PI3K-mediated inhibition and its effects on Aurora B and downstream mediators AKT and PDK1. * indicates components that are important for survival against PI3K inhibition. Cl, cleaved; Emp, empty vector; NS, nonspecific siRNA.
Because total AKT levels were upregulated upon Aurora B overexpression in NOTCH1MUT HNSCC cells with concordant protection from PI3K inhibition-induced apoptosis, we examined the effect of total AKT on PI3K inhibition-induced apoptosis. We manipulated AKT1, the predominant isoform of AKT. Overexpression of AKT1 in NOTCH1MUT HNSCC cells significantly reduced PI3K inhibition-induced apoptosis (Fig. 5E,F). Moreover, we observed corresponding changes in the PDK1 protein levels but not Aurora B levels upon AKT1 overexpression in NOTCH1MUT HNSCC cells (Fig. 5E). Additionally, AKT1 knockdown in NOTCH1WT HNSCC cells treated with PI3K inhibition resulted in markedly higher cell death (Fig. 5G,H, Supplementary Fig. S7C,D).
These findings illustrate that total Aurora B governs the expression of total AKT, which subsequently regulates PDK1 levels in NOTCH1MUT HNSCC cells, in which Aurora B, AKT, and PDK1 are crucial effectors that determine cell survival in response to PI3K inhibition. However, in NOTCH1WT HNSCC cells, Aurora B does not regulate total AKT or PDK1 levels (Fig. 5I).
DISCUSSION
To address the need for biomarker-selected targeted therapy for HNSCC, we previously demonstrated that PI3K inhibition caused apoptosis selectively in NOTCH1MUT HNSCC, but the mechanisms of resistance in NOTCH1WT HNSCC were unknown. In this study, we show that PI3K inhibition leads to reduced Aurora B levels, which in turn regulate total AKT protein levels exclusively in NOTCH1MUT HNSCC in a kinase-independent manner. Subsequently, AKT affects PDK1 protein levels, also independent of kinase activity. Total AKT and PDK1 loss mediates apoptosis after PI3K inhibition in NOTCH1MUT HNSCC. In addition, the pathways involving maintenance of protein levels of Aurora kinases in response to PI3K inhibition contribute to both innate resistance and AR to PI3K inhibition in HNSCC. In our study, concurrent Aurora kinase and PI3K inhibition led to increased cell death in vitro and in vivo. In contrast, reduced Aurora A and FOXM1 levels were associated with, but did not control the apoptotic phenotype in NOTCH1MUT HNSCC cells.
The mechanism we propose identifies several previously unrecognized interactions between the PI3K/AKT pathway and Aurora kinases. Our discovery raises several questions for future studies that would dissect specific interactions within this pathway, including understanding how Aurora B levels are differentially regulated in NOTCH1WT and NOTCH1MUT HNSCC in response to PI3K inhibition. Because Aurora B levels are altered at both mRNA and protein levels, it is important to determine the molecular factors responsible for these changes. One potential mediator may be FOXO3a. We observed significantly lower levels of phosphorylated FOXO3a in response to PI3K inhibition in NOTCH1MUT HNSCC cells than in NOTCH1WT cells. FOXO3a undergoes AKT-mediated phosphorylation at S235 and is rendered inactive, thus being unavailable to bind to the promoters of its numerous targets, including Aurora B, and repress their transcription (35). Involvement of FOXO3a may contribute to the regulation of AURKB mRNA levels. For identification of posttranscriptional regulators of Aurora B, it will be imperative to determine the protein half-life of Aurora B in NOTCH1WT and NOTCH1MUT HNSCC cells with and without PI3K inhibition.
Another striking finding from the current study is the sustained, depleted protein levels of total AKT in NOTCH1MUT HNSCC in response to PI3K inhibition. Unlike Aurora B, AKT is clearly regulated at the posttranscriptional level. Moreover, the effect of Aurora B protein expression on AKT is not solely dependent upon kinase activity. Therefore, it will be intriguing to investigate how Aurora B and AKT interact with each other in a kinase-independent manner. Furthermore, our time course studies showed that AKT protein levels initially decrease in NOTCH1WT and NOTCH1MUT cells with more marked, durable changes in NOTCH1MUT. Further work is warranted to understand the mechanism behind AKT protein downregulation. One potential candidate to mediate this differential effect is BRCA1. BRCA1 mutant cells accumulate nuclear phospho-AKT and consequently inactivate the transcription functions of FOXO3a, a main nuclear target of phospho-AKT (36). Additionally, NOTCH1 activation further compensates for BRCA1 deficiency and promotes survival of triple-negative breast cancer (37). Furthermore, BRCA1 phosphorylation is regulated by the PI3K pathway, and thereby its subcellular localization and functions (38). In summary, the differential sensitivity to PI3K inhibition in NOTCH1WT and NOTCH1MUT HNSCC could be due to the differential activation of BRCA1, which could also explain the proteomic alterations in AKT and Aurora B (39).
Another aspect that remains to be understood is how total AKT1 regulates PDK1 levels in NOTCH1MUT but not in NOTCH1WT HNSCC. A recent study showed that CK1- and GSK3β-mediated phosphorylation of PDK1 led to its ubiquitination and degradation by E3 ubiquitin ligase speckle type BTB/POZ protein (SPOP) (40). We speculate that increased AKT levels lead to inactivation of GSK3β, which then fails to phosphorylate PDK1, leading to its degradation by SPOP. It is possible that knockdown of AKT1 alone in NOTCH1WT HNSCC does not alter PDK1 to a significant extent by itself, because the other AKT isoforms could function as redundant proteins in this context. However, when the PI3K pathway is inhibited in AKT1 knockdown cells, activated GSK3β could potentially mediate PI3K degradation in NOTCH1WT HNSCC. This mechanism could potentially explain our observation of differential PDK1 levels upon AKT1 manipulation in NOTCH1MUT and NOTCH1WT HNSCC in response to PI3K inhibition.
We showed that the combination of PI3K and Aurora kinase inhibition is synergistic and leads to increased apoptosis in most HNSCC cells independent of mutation status, including those with innate and AR to PI3K inhibitors. These in vitro findings were validated in our in vivo models, which showed robust tumor regression in mice receiving combined therapy. Our findings suggest that this combination would be broadly effective against HNSCC in patients who may have heterogeneous tumors. In addition, it is rational to target a pathway that mediates AR initially to achieve a more durable response to therapy (41).
The mechanism that underlies the synergy between PI3K and Aurora kinase inhibition is likely distinct from the model we propose to explain the resistance of NOTCH1WT HNSCC to PI3K inhibitors, which is independent of Aurora A and the kinase activity of Aurora B. One possible explanation for the synergy is that because PI3K inhibition leads to reduced total levels of Aurora kinases, these cells are more dependent on the remaining activity of Aurora kinases for mitotic progression. Additionally, prolonged inhibition of Aurora A can lead to inhibition of Aurora B (27). A second possible explanation for the synergy hinges on the finding that PI3K inhibition leads to decreased Rb protein expression in both NOTCH1MUT and NOTCH1WT HNSCC cell lines. Two independent studies have shown that cancer cells with loss of RB1 are hyper-dependent on Aurora A and Aurora B for survival (42, 43).
Donnella, et al. demonstrated a decrease in total AURKA mRNA and protein 24 hours following PI3K inhibition in sensitive breast cancer cell lines (34). The combination of a PI3K inhibitor with alisertib was synergistic in 38% of breast cancer lines. They demonstrated that MYC-driven AURKA expression maintains AKT and mTOR activity; inhibition of Aurora A enhances PI3K inhibition by contributing to the complete suppression of AKT/mTOR signaling. In contrast to our model in HNSCC, they found that AURKB was not significantly associated with sensitivity to PI3K inhibitors in breast cancer.
The mechanisms of AR to PI3K inhibition in NOTCH1MUT HNSCC could be driven by additional mechanisms that do not overlap with mechanisms of innate resistance in NOTCH1WT HNSCC. As is the case with targeted therapies in non-small cell lung cancer, there are several distinct mechanisms of AR in epidermal growth factor (EGFR) mutant and anaplastic lymphoma kinase (ALK) positive subsets. They either involve on-target mechanisms involving gene/target amplification that enables continuous downstream signaling or off-target effects which results in activation of bypass signaling (44).
Because both PI3K (e.g., paxalisib, umbralisib, parsaclisib, copanlisib, and duvelisib) and Aurora kinase inhibitors (alisertib, barasertib) are in clinical development, our work could be rapidly translated to clinical testing. Alternatively, AKT and Aurora kinase-specific proteolysis-targeting chimeras (PROTACS), which are in the process of development and validation, might be an effective therapeutic option in cases where PI3K inhibitors fail (25, 45, 46). Notably, our recent clinical trial testing a dual PI3K/mTOR inhibitor in NOTCH1MUT HNSCC patients with recurrent or metastatic disease showed modest single-agent clinical activity (NCT03740100), indicating that combination therapy could be an effective approach (8, 47).
These findings collectively show that sustained Aurora B expression via AKT and PDK1 levels drives resistance to PI3K inhibition-induced apoptosis in NOTCH1WT HNSCC. We have defined a mechanism that drives sensitivity and resistance to PI3K inhibitors in NOTCH1MUT HNSCC and propose combined PI3K and Aurora kinase inhibition to maximize clinical efficacy and overcome innate and AR to PI3K inhibitors, thereby establishing a foundation for future clinical trials.
Supplementary Material
Statement of significance:
Aurora B signaling facilitates resistance to PI3K inhibition in head and neck squamous cell carcinoma, suggesting that combined inhibition of PI3K and Aurora kinase is a rational therapeutic strategy to overcome resistance.
Acknowledgments:
We thank Erica Goodoff in MD Anderson’s Research Medical Library for editing the manuscript. This work was supported by philanthropic contributions to The University of Texas MD Anderson Cancer Center’s Oropharynx Discovery Program (to FMJ), from the National Institutes of Health (1R01CA235620, to FMJ, MJF), and the Cancer Prevention and Research Institute of Texas (RP200369, to FMJ, MJF). Flow cytometry and bioinformatics analyses were supported by the National Cancer Institute through MD Anderson’s Cancer Center Support Grant (P30CA016672).
Footnotes
Competing interests: FMJ has received research funding from PIQUR, Takeda, and Trovagene. Other authors have no conflicts of interest to declare.
References and Notes
- 1.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, Zhang N, El-Naggar AK, Jasser SA, Weinstein JN, Trevino L, Drummond JA, Muzny DM, Wu Y, Wood LD, Hruban RH, Westra WH, Koch WM, Califano JA, Gibbs RA, Sidransky D, Vogelstein B, Velculescu VE, Papadopoulos N, Wheeler DA, Kinzler KW, Myers JN, Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 333, 1154–1157 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pickering CR, Zhang J, Yoo SY, Bengtsson L, Moorthy S, Neskey DM, Zhao M, Ortega Alves MV, Chang K, Drummond J, Cortez E, Xie TX, Zhang D, Chung W, Issa JP, Zweidler-McKay PA, Wu X, El-Naggar AK, Weinstein JN, Wang J, Muzny DM, Gibbs RA, Wheeler DA, Myers JN, Frederick MJ, Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov 3, 770–781 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.N. Cancer Genome Atlas, Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortes ML, Auclair D, Berger MF, Saksena G, Guiducci C, Onofrio RC, Parkin M, Romkes M, Weissfeld JL, Seethala RR, Wang L, Rangel-Escareno C, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Winckler W, Ardlie K, Gabriel SB, Meyerson M, Lander ES, Getz G, Golub TR, Garraway LA, Grandis JR, The mutational landscape of head and neck squamous cell carcinoma. Science 333, 1157–1160 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shah PA, Huang C, Li Q, Kazi SA, Byers LA, Wang J, Johnson FM, Frederick MJ, NOTCH1 Signaling in Head and Neck Squamous Cell Carcinoma. Cells 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sambandam V, Frederick MJ, Shen L, Tong P, Rao X, Peng S, Singh R, Mazumdar T, Huang C, Li Q, Pickering CR, Myers JN, Wang J, Johnson FM, PDK1 Mediates NOTCH1-Mutated Head and Neck Squamous Carcinoma Vulnerability to Therapeutic PI3K/mTOR Inhibition. Clin Cancer Res 25, 3329–3340 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazumdar T, Byers LA, Ng PK, Mills GB, Peng S, Diao L, Fan YH, Stemke-Hale K, Heymach JV, Myers JN, Glisson BS, Johnson FM, A comprehensive evaluation of biomarkers predictive of response to PI3K inhibitors and of resistance mechanisms in head and neck squamous cell carcinoma. Mol Cancer Ther 13, 2738–2750 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson FM, Janku F, Lee JJ, Schmitz D, Streefkerk H, Frederick M, Single-arm study of bimiralisib in head and neck squamous cell carcinoma (HNSCC) patients (pts) harboring NOTCH1 loss of function (LOF) mutations. J Clin Oncol 38, (2020). [Google Scholar]
- 9.Pickering CR, Zhou JH, Lee JJ, Drummond JA, Peng SA, Saade RE, Tsai KY, Curry JL, Tetzlaff MT, Lai SY, Yu J, Muzny DM, Doddapaneni H, Shinbrot E, Covington KR, Zhang J, Seth S, Caulin C, Clayman GL, El-Naggar AK, Gibbs RA, Weber RS, Myers JN, Wheeler DA, Frederick MJ, Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin Cancer Res 20, 6582–6592 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agrawal N, Jiao Y, Bettegowda C, Hutfless SM, Wang Y, David S, Cheng Y, Twaddell WS, Latt NL, Shin EJ, Wang LD, Wang L, Yang W, Velculescu VE, Vogelstein B, Papadopoulos N, Kinzler KW, Meltzer SJ, Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov 2, 899–905 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martincorena I, Fowler JC, Wabik A, Lawson ARJ, Abascal F, Hall MWJ, Cagan A, Murai K, Mahbubani K, Stratton MR, Fitzgerald RC, Handford PA, Campbell PJ, Saeb-Parsy K, Jones PH, Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.N. Cancer Genome Atlas Research, Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalu NN, Mazumdar T, Peng S, Shen L, Sambandam V, Rao X, Xi Y, Li L, Qi Y, Gleber-Netto FO, Patel A, Wang J, Frederick MJ, Myers JN, Pickering CR, Johnson FM, Genomic characterization of human papillomavirus-positive and -negative human squamous cell cancer cell lines. Oncotarget 8, 86369–86383 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalu NN, Mazumdar T, Peng S, Tong P, Shen L, Wang J, Banerjee U, Myers JN, Pickering CR, Brunell D, Stephan CC, Johnson FM, Comprehensive pharmacogenomic profiling of human papillomavirus-positive and -negative squamous cell carcinoma identifies sensitivity to aurora kinase inhibition in KMT2D mutants. Cancer Lett 431, 64–72 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Zhang M, Singh R, Peng S, Mazumdar T, Sambandam V, Shen L, Tong P, Li L, Kalu NN, Pickering CR, Frederick M, Myers JN, Wang J, Johnson FM, Mutations of the LIM protein AJUBA mediate sensitivity of head and neck squamous cell carcinoma to treatment with cell-cycle inhibitors. Cancer Lett 392, 71–82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrarotto R, Goonatilake R, Yoo SY, Tong P, Giri U, Peng S, Minna J, Girard L, Wang Y, Wang L, Li L, Diao L, Peng DH, Gibbons DL, Glisson BS, Heymach JV, Wang J, Byers LA, Johnson FM, Epithelial-Mesenchymal Transition Predicts Polo-Like Kinase 1 Inhibitor-Mediated Apoptosis in Non-Small Cell Lung Cancer. Clin Cancer Res 22, 1674–1686 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J, He X, Baggerly KA, Coombes KR, Hennessy BT, Mills GB, Non-parametric quantification of protein lysate arrays. Bioinformatics 23, 1986–1994 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Zumsteg ZS, Morse N, Krigsfeld G, Gupta G, Higginson DS, Lee NY, Morris L, Ganly I, Shiao SL, Powell SN, Chung CH, Scaltriti M, Baselga J, Taselisib (GDC-0032), a Potent beta-Sparing Small Molecule Inhibitor of PI3K, Radiosensitizes Head and Neck Squamous Carcinomas Containing Activating PIK3CA Alterations. Clin Cancer Res 22, 2009–2019 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chou TC, Talalay P, Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 22, 27–55 (1984). [DOI] [PubMed] [Google Scholar]
- 20.Chou TC, The median-effect principle and the combination index for quantitation of synergism and antagonism. Chou TC, Rideout DC, Eds., Synergism and Antagonism in Chemotherapy (Academic Press, San Diego, CA, 1991), pp. 61–102. [Google Scholar]
- 21.Chou TC, Riedeout D, Chou J, Bertino JR, Dulbecco R, Chemotherapeutic synergism, potential and antagonism. Dulbecco R, Ed., Encyclopedia of Human Biology (Academic Press, San Diego, CA, 1991), vol. 2, pp. 371–379. [Google Scholar]
- 22.Benjamini Y, Hochberg Y, Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Statistical Methodology 57, 289–300 (1995). [Google Scholar]
- 23.Munster P, Aggarwal R, Hong D, Schellens JH, van der Noll R, Specht J, Witteveen PO, Werner TL, Dees EC, Bergsland E, Agarwal N, Kleha JF, Durante M, Adams L, Smith DA, Lampkin TA, Morris SR, Kurzrock R, First-in-Human Phase I Study of GSK2126458, an Oral Pan-Class I Phosphatidylinositol-3-Kinase Inhibitor, in Patients with Advanced Solid Tumor Malignancies. Clin Cancer Res 22, 1932–1939 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Kunitoku N, Sasayama T, Marumoto T, Zhang D, Honda S, Kobayashi O, Hatakeyama K, Ushio Y, Saya H, Hirota T, CENP-A phosphorylation by Aurora-A in prophase is required for enrichment of Aurora-B at inner centromeres and for kinetochore function. Dev Cell 5, 853–864 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Xu J, Yu X, Martin TC, Bansal A, Cheung K, Lubin A, Stratikopoulos E, Cahuzac KM, Wang L, Xie L, Zhou R, Shen Y, Wu X, Yao S, Qiao R, Poulikakos PI, Chen X, Liu J, Jin J, Parsons R, AKT Degradation Selectively Inhibits the Growth of PI3K/PTEN Pathway–Mutant Cancers with Wild-Type KRAS and BRAF by Destabilizing Aurora Kinase B. Cancer Discovery 11, 3064–3089 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janku F, Yap TA, Meric-Bernstam F, Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol 15, 273–291 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Martin D, Fallaha S, Proctor M, Stevenson A, Perrin L, McMillan N, Gabrielli B, Inhibition of Aurora A and Aurora B Is Required for the Sensitivity of HPV-Driven Cervical Cancers to Aurora Kinase Inhibitors. Mol Cancer Ther 16, 1934–1941 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Schwartz GK, Carvajal RD, Midgley R, Rodig SJ, Stockman PK, Ataman O, Wilson D, Das S, Shapiro GI, Phase I study of barasertib (AZD1152), a selective inhibitor of Aurora B kinase, in patients with advanced solid tumors. Invest New Drugs 31, 370–380 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Mortlock AA, Foote KM, Heron NM, Jung FH, Pasquet G, Lohmann JJ, Warin N, Renaud F, De Savi C, Roberts NJ, Johnson T, Dousson CB, Hill GB, Perkins D, Hatter G, Wilkinson RW, Wedge SR, Heaton SP, Odedra R, Keen NJ, Crafter C, Brown E, Thompson K, Brightwell S, Khatri L, Brady MC, Kearney S, McKillop D, Rhead S, Parry T, Green S, Discovery, synthesis, and in vivo activity of a new class of pyrazoloquinazolines as selective inhibitors of aurora B kinase. J Med Chem 50, 2213–2224 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, Tan Y, Ackerson T, Costa RH, Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol 25, 10875–10894 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Yun Y, Wu B, Wen L, Wen M, Yang H, Zhao L, Liu W, Huang S, Wen N, Li Y, FOXM1 promotes reprogramming of glucose metabolism in epithelial ovarian cancer cells via activation of GLUT1 and HK2 transcription. Oncotarget 7, 47985–47997 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui J, Shi M, Xie D, Wei D, Jia Z, Zheng S, Gao Y, Huang S, Xie K, FOXM1 promotes the warburg effect and pancreatic cancer progression via transactivation of LDHA expression. Clin Cancer Res 20, 2595–2606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park HJ, Carr JR, Wang Z, Nogueira V, Hay N, Tyner AL, Lau LF, Costa RH, Raychaudhuri P, FoxM1, a critical regulator of oxidative stress during oncogenesis. EMBO J 28, 2908–2918 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donnella HJ, Webber JT, Levin RS, Camarda R, Momcilovic O, Bayani N, Shah KN, Korkola JE, Shokat KM, Goga A, Gordan JD, Bandyopadhyay S, Kinome rewiring reveals AURKA limits PI3K-pathway inhibitor efficacy in breast cancer. Nat Chem Biol 14, 768–777 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osei-Sarfo K, Gudas LJ, Retinoids induce antagonism between FOXO3A and FOXM1 transcription factors in human oral squamous cell carcinoma (OSCC) cells. PLoS One 14, e0215234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang T, Ohashi A, Huang Y, Pandita TK, Ludwig T, Powell SN, Yang Q, Negative Regulation of AKT Activation by BRCA1. Cancer Res 68, 10040–10044 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miao K, Lei JH, Valecha MV, Zhang A, Xu J, Wang L, Lyu X, Chen S, Miao Z, Zhang X, Su SM, Shao F, Rajendran BK, Bao J, Zeng J, Sun H, Chen P, Tan K, Chen Q, Wong KH, Xu X, Deng CX, NOTCH1 activation compensates BRCA1 deficiency and promotes triple-negative breast cancer formation. Nat Commun 11, 3256 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma B, Guo W, Shan M, Zhang N, Ma B, Sun G, BRCA1 subcellular localization regulated by PI3K signaling pathway in triple-negative breast cancer MDA-MB-231 cells and hormone-sensitive T47D cells. Open Life Sci 15, 501–510 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryser S, Dizin E, Jefford CE, Delaval B, Gagos S, Christodoulidou A, Krause KH, Birnbaum D, Irminger-Finger I, Distinct roles of BARD1 isoforms in mitosis: full-length BARD1 mediates Aurora B degradation, cancer-associated BARD1beta scaffolds Aurora B and BRCA2. Cancer Res 69, 1125–1134 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Jiang Q, Zheng N, Bu L, Zhang X, Zhang X, Wu Y, Su Y, Wang L, Zhang X, Ren S, Dai X, Wu D, Xie W, Wei W, Zhu Y, Guo J, SPOP-mediated ubiquitination and degradation of PDK1 suppresses AKT kinase activity and oncogenic functions. Mol Cancer 20, 100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neel DS, Bivona TG, Resistance is futile: overcoming resistance to targeted therapies in lung adenocarcinoma. npj Precision Oncology 1, 3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oser MG, Fonseca R, Chakraborty AA, Brough R, Spektor A, Jennings RB, Flaifel A, Novak JS, Gulati A, Buss E, Younger ST, McBrayer SK, Cowley GS, Bonal DM, Nguyen QD, Brulle-Soumare L, Taylor P, Cairo S, Ryan CJ, Pease EJ, Maratea K, Travers J, Root DE, Signoretti S, Pellman D, Ashton S, Lord CJ, Barry ST, Kaelin WG Jr., Cells Lacking the RB1 Tumor Suppressor Gene Are Hyperdependent on Aurora B Kinase for Survival. Cancer Discov 9, 230–247 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong X, Du J, Parsons SH, Merzoug FF, Webster Y, Iversen PW, Chio LC, Van Horn RD, Lin X, Blosser W, Han B, Jin S, Yao S, Bian H, Ficklin C, Fan L, Kapoor A, Antonysamy S, Mc Nulty AM, Froning K, Manglicmot D, Pustilnik A, Weichert K, Wasserman SR, Dowless M, Marugan C, Baquero C, Lallena MJ, Eastman SW, Hui YH, Dieter MZ, Doman T, Chu S, Qian HR, Ye XS, Barda DA, Plowman GD, Reinhard C, Campbell RM, Henry JR, Buchanan SG, Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of the RB1 Tumor Suppressor Gene. Cancer Discov 9, 248–263 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Lovly CM, Iyengar P, Gainor JF, Managing Resistance to EFGR- and ALK-Targeted Therapies. Am Soc Clin Oncol Educ Book 37, 607–618 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.You I, Erickson EC, Donovan KA, Eleuteri NA, Fischer ES, Gray NS, Toker A, Discovery of an AKT Degrader with Prolonged Inhibition of Downstream Signaling. Cell Chem Biol 27, 66–73 e67 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adhikari B, Bozilovic J, Diebold M, Schwarz JD, Hofstetter J, Schroder M, Wanior M, Narain A, Vogt M, Dudvarski Stankovic N, Baluapuri A, Schonemann L, Eing L, Bhandare P, Kuster B, Schlosser A, Heinzlmeir S, Sotriffer C, Knapp S, Wolf E, PROTAC-mediated degradation reveals a non-catalytic function of AURORA-A kinase. Nat Chem Biol 16, 1179–1188 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Janku F, Johnson FM, Opyrchal M, Dowlati A, Hierro C, Forester M, Blagden SP, Wicki A, Schmitz D, Adjei AA, Abstract B109: Oral dual PI3K/mTOR inhibitor bimiralisib demonstrates tolerability and a signal of activity in head and neck squamous cell cancer with NOTCH1 loss-of-function mutation. Molecular Cancer Therapeutics 18, B109–B109 (2019). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its supplementary data files. Derived data supporting the findings of this study are available from the corresponding author upon request.