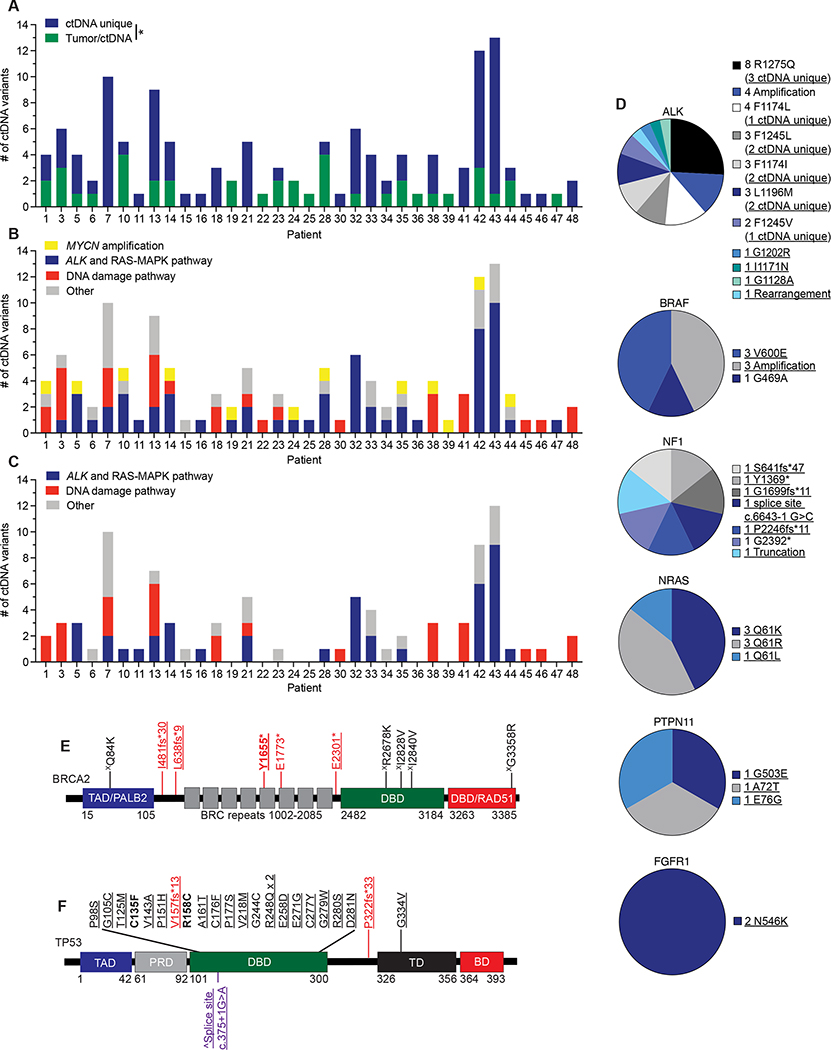
Figure 3. ctDNA unique pathogenic variants in cancer driver genes are commonly identified in relapsed neuroblastoma patients.
(A) Plot of number of ctDNA unique versus ctDNA/tumor common variants for evaluable patients.
(B) Plot of major categories of all ctDNA variants identified.
(C) Plot of major categories of ctDNA unique variants identified.
(D) Charts of ctDNA defined alterations in ALK and RAS-MAPK pathway genes.
(E, F) ctDNA identified genetic variants in BRCA2 (E) and TP53 (F) transposed on their respective protein domains.
Underlined variants denote those unique to ctDNA and (X) denotes variants of unknown significance. In E and F, bolded variants represent those suspected to be of germline origin; red, denotes putative loss-of-function (LOF) variants; purple, denotes variants in splice sites; black, denotes missense variants. (^) denotes splice site variant in TP53 that is predicted to lead to a truncated protein (57). Relative protein domains in E and F (not drawn to scale) (58–60); TAD, transactivation domain; PALB2, PALB2 binding domain; DBD, DNA binding domain; RAD51, RAD51 binding domain; PRD, proline-rich domain; TD, tetramerization domain; BD; basic domain.
*, p<0.05.