Figure 7. ctDNA-identified ERRFI1 variants are pathogenic in neuroblastoma cells.
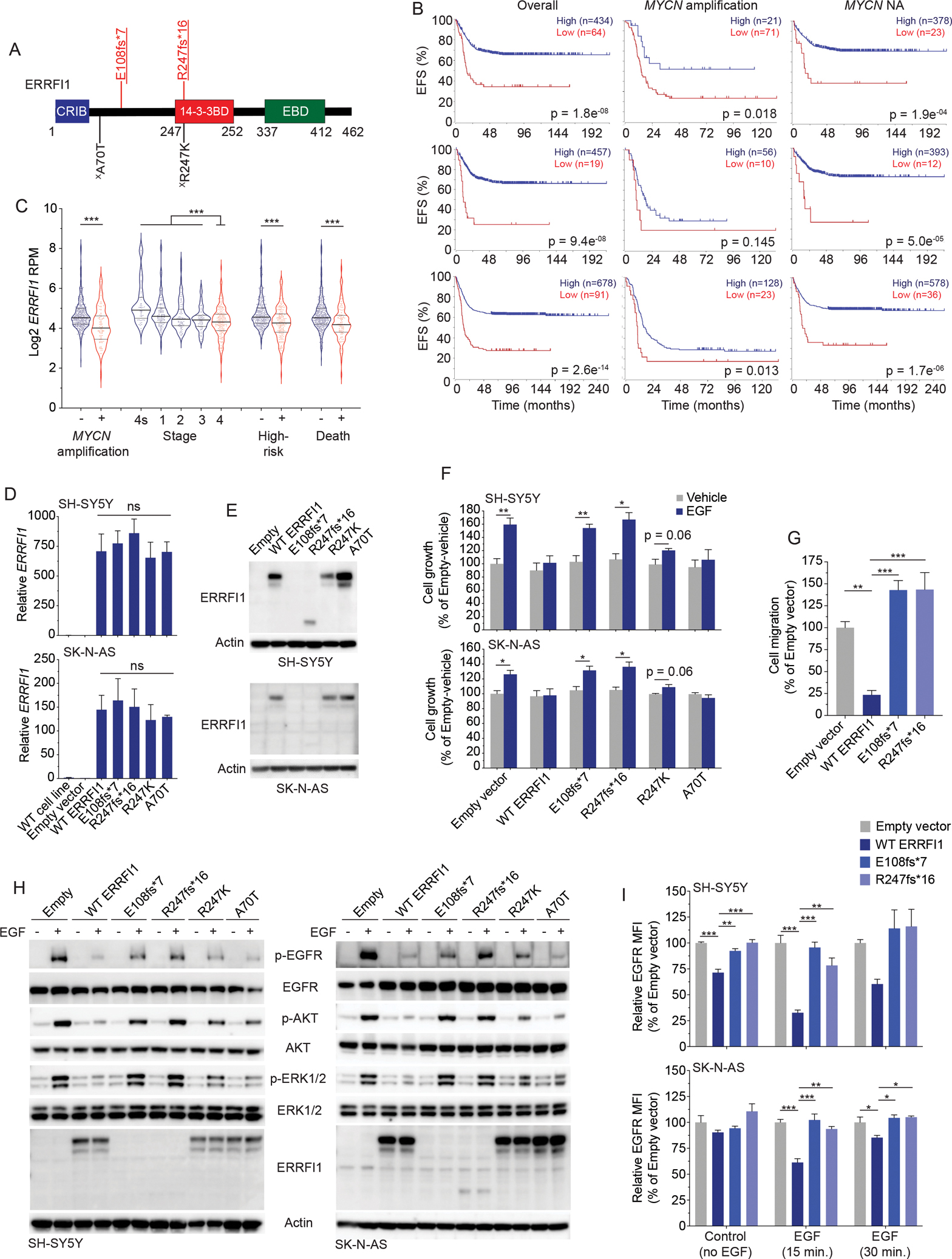
(A) ctDNA-identified variants in ERRFI1 transposed on the ERRFI1 protein domains.
(B) Neuroblastoma event-free survival (EFS) plots stratified by ERRFI1 expression for all patients (left), patients with MYCN amplified tumors (middle), and patients with MYCN non-amplified (NA) tumors (right) in 3 large neuroblastoma data sets, SEQC (n=498, top), Kocak (n=649; middle), and Cangelosi (n=786; bottom). EFS plots were generated in the Genomics Analysis and Visualization Platform (R2; https://hgserver1.amc.nl/cgi-bin/r2/main.cgi).
(C) ERRFI1 expression in neuroblastoma tumors stratified by several clinical covariates (SEQC; n=498 tumors).
(D, E) ERRFI1 expression in ERRFI1 isogenic SH-SY5Y and SK-N-AS neuroblastoma cell lines by RT-PCR (D) and western blot (E).
(F) Relative cell growth plots of ERRFI1 isogenic SH-SY5Y (top) and SK-N-AS (bottom) neuroblastoma cell lines after EGF stimulation (50 ng/mL).
(G) Relative EGF-induced cell migration of ERRFI1 isogenic SK-N-AS neuroblastoma cells.
(H) Western blot of ERRFI1 isogenic SH-SY5Y (left) and SK-N-AS (right) neuroblastoma cell lines after EGF stimulation (50 ng/mL).
(I) Relative EGFR cell surface levels of ERRFI1 isogenic SH-SY5Y (top) and SK-N-AS (bottom) neuroblastoma cell lines with and without EGF stimulation (50 ng/mL).
Underlined variants denote those unique to ctDNA and (X) denotes variants of unknown significance. In A, red denotes putative LOF variant and black denotes missense VUS. Relative protein domains in A (not drawn to scale); CRIB, Cdc42/Rac interactive binding domain; 14–3-3BD, 14–3-3 binding domain, EBD, EGFR or ErbB binding domain.
WT, wild type.
*, p<0.05; **, p<0.01; ***, p<0.001; ns, not significant.
