Abstract
Autophagy is a conserved catabolic process that maintains cellular homeostasis. Autophagy supports lung tumorigenesis and is a potential therapeutic target in lung cancer. A better understanding of the importance of tumor cell-autonomous versus systemic autophagy in lung cancer could facilitate clinical translation of autophagy inhibition. Here, we exploited inducible expression of Atg5 shRNA to temporally control Atg5 levels and generate reversible tumor-specific and systemic autophagy loss mouse models of KrasG12D/+;p53−/− (KP) non-small cell lung cancer (NSCLC). Transient suppression of systemic but not tumor Atg5 expression significantly reduced established KP lung tumor growth without damaging normal tissues. In vivo 13C isotope tracing and metabolic flux analyses demonstrated that systemic Atg5 knockdown specifically led to reduced glucose and lactate uptake. As a result, carbon flux from glucose and lactate to major metabolic pathways, including the tricarboxylic acid cycle, glycolysis, and serine biosynthesis, was significantly reduced in KP NSCLC following systemic autophagy loss. Furthermore, systemic Atg5 knockdown increased tumor T cell infiltration, leading to T cell-mediated tumor killing. Importantly, intermittent transient systemic Atg5 knockdown, which resembles what would occur during autophagy inhibition for cancer therapy, significantly prolonged lifespan of KP lung tumor-bearing mice, resulting in recovery of normal tissues but not tumors. Thus, systemic autophagy supports the growth of established lung tumors by promoting immune evasion and sustaining cancer cell metabolism for energy production and biosynthesis, and the inability of tumors to recover from loss of autophagy provides further proof of concept that inhibition of autophagy is a valid approach to cancer therapy.
Keywords: autophagy, Kras, p53, lung tumor, immune evasion, cancer metabolism
Introduction
Lung cancer is the leading cause of cancer-related death worldwide, with Non-small cell lung cancer (NSCLC) accounting for more than 85% of these cases (1). Oncogenic co-mutations of TP53 and KRAS (either KRASG12D or KRASG12C) (KP) represents one subtype of KRAS-driven lung cancer, accounting for 15-20% of NSCLC with distinct biological and therapeutic vulnerabilities. With the recent exception of KRASG12C, targeting oncogenic RAS directly has not been effective, and inhibiting downstream effectors of RAS signaling such as the MAP kinase pathway has not produced durable responses (2-4). Immunotherapy with pembrolizumab and nivolumab (humanized monoclonal anti-PD-1 antibodies), and atezolizumab and durvalumab (humanized monoclonal anti-PD-L1 antibodies) have been approved for lung cancer treatment (5,6). However, only 19.4% of NSCLC patients treated with single-agent Pembrolizumab have exhibited an objective response and a median duration of response is only 12.5 months (7). Among them, patients with co-mutations of KRAS and TP53 showed better response, which could be due to the significantly increased expression of immune checkpoints such as PD-L1, activated T-effector cell and interferon-γ, and higher mutation burdens in p53 mutant tumors compared with p53 wild type (WT) tumors (8-10). Nonetheless, most patients with KP mutations do not benefit from immunotherapy. Recently, KRASG12C inhibitors have led to a substantial reduction in mortality in the treatment of KRAS-mutant NSCLC. Responses to KRASG12C inhibitors are observed across the range of TP53, LKB1 and KEAP1 co-occurring mutations. Unfortunately, approximately 40% of patients do not respond or acquire resistance to the treatment, which limits the clinical benefit of KRASG12C inhibitors (11-15). Therefore, new treatments are still urgently needed for KRAS mutant NSCLC.
KRAS activation rewires cancer cell metabolism for rapid proliferation, including upregulation of glucose and glutamine uptake, promotion of micropinocytosis, and activation of autophagy to sustain metabolism, biosynthesis, redox homeostasis, and survival (16). Targeting cancer metabolism is already a successful approach to cancer treatment and recent advances in our understanding of tumor metabolism are revealing new targets (17-20). Autophagy degrades cellular waste, such as protein aggregates and damaged organelles, and recycles their building blocks to maintain cellular homeostasis (21,22). Through the analysis of systemic and tissue-specific essential autophagy gene (Atg) knockout mice, it has been shown that autophagy plays a critical role in mammalian development, differentiation, and disease (23). Loss of autophagy in humans (24) and adult mice is associated with the onset of various diseases, such as neurodegeneration, and liver inflammation (25-31). In contrast, oncogenic RAS activation renders tumor cells addicted to autophagy enabling survival to metabolic stress (32-34). Thus, there are examples where both autophagy loss and activation contribute to disease.
Using genetically engineered mouse models (GEMMs) to conditionally delete Atgs, our group and others have demonstrated that autophagy within both tumor cells and the host supports tumorigenesis in different types of cancer via distinct mechanisms. Autophagy within tumor cells promotes tumorigenesis by inhibiting p53 activation, sustaining redox homeostasis, maintaining essential amino acid levels for energy production and biosynthesis, and inhibiting antigen presentation (35-42). Autophagy within the host promotes tumorigenesis by maintaining circulating arginine (43) and alanine and other amino acids in the tumor microenvironment (42,44), and by suppressing an anti-tumor T-cell response through autophagy-mediated “hepatic autophagy immune tolerance” (45). Thus, autophagy promotes cancer by enhancing metabolic fitness and limiting immune and non-immune tumor suppression mechanisms (22,46), providing strong evidence that targeting the autophagy pathway with small molecule inhibitors could be a novel therapeutic strategy. Indeed, recent preclinical mouse studies and clinical trials suggest that blocking autophagy by targeting lysosome function using hydroxychloroquine (HCQ) can increase sensitivity to immunotherapy or a MAP kinase pathway inhibition downstream of oncogenic RAS in cancer treatment (35,47,48).
Using the KrasLSL-G12D/+;p53Flox/Flox;Atg7Flox/Flox GEMM for KrasG12D/+;p53−/− (KP) NSCLC, we previously found that autophagy supports KP lung tumorigenesis (36). However, the deletion of Atg7 occurs simultaneously with activation of the oncogenic drivers, which cannot distinguish whether switching off autophagy in established tumors would have anti-tumor activity. We therefore generated a whole body conditional Atg7 knockout GEMM for KP NSCLC and found that conditional Atg7 deletion in the tumor and host impaired established KP lung tumor growth (49). In these whole-body conditional Atg7 knockout mice, loss of autophagy is irreversible and although there was marked tumor regression within 4-5 weeks, mice die within 2-3 months predominantly due to neurodegeneration (49). While informatively demonstrating that tumors are more sensitive to loss of autophagy than most normal tissues, the irreversibility of autophagy deficiency does not fully simulate the situation of autophagy inhibition in clinical practice. In addition, the damage to liver, pancreas and muscle by host autophagy ablation was observed (49-51). Therefore, understanding whether the damage to normal and tumor tissues caused by autophagy ablation is reversible after autophagy restoration is of great significance for translating autophagy inhibition to treat KP NSCLC.
In this study, we generated an autophagy loss switchable GEMMs for KP NSCLC by crossing an inducible shRNA mouse model that leads to temporal control of Atg5 levels ubiquitously or cell type-specifically (50,51) with KrasLSL-G12D/+;p53Flox/Flox mice to study the role of tumor cell-autonomous and systemic autophagy in established KP lung tumors. We found that systemic Atg5 knockdown, but not tumor-specific Atg5 knockdown, significantly inhibited KP lung tumor growth. We further explored the underlying mechanisms by which systemic autophagy supports established KP lung tumor growth. We found that systemic Atg5 knockdown reduced glucose and lactate uptake in KP lung tumors, leading to decreased carbon flux from glucose to TCA cycle intermediates and serine. In addition, we observed that KP lung tumors hijack truncated gluconeogenesis for biosynthesis, which was impaired by systemic Atg5 knockdown. Moreover, we observed that systemic Atg5 knockdown significantly increased tumor-infiltrating lymphocytes and supported T cell-mediated tumor killing. Most importantly, the impaired tumor gluconeogenesis and biosynthesis and increased tumor CD8 T cell infiltration were sustained even when Atg5 expression was restored for a short term. Finally, in stark contrast with normal tissues, the reduction of tumor growth by Atg5 knockdown was irreversible when systemic autophagy is restored in the short term. Cycling transient systemic Atg5 knockdown and restoration in KP lung tumor bearing mice significantly prolonged mouse lifespan, resulting in recovery of normal tissues. Taken together, our findings reinforce the concept that systemically targeting autophagy may be a potential cancer therapeutic strategy.
Methods
Mice
Rutgers Animal Care and Use Committee (IACUC) has approved all animal experiments performed in this study. Transgenic mouse line with Tet-regulated shRNA expression against essential autophagy gene Atg5 that enables temporal control of Atg5 levels cell type-specifically (LSL-rtTA3;shRNA-Atg5) (51) was bred with KrasLSL-G12D/+;p53Flox/Flox GEMM to obtain the tumor-specific autophagy inducible LSL-rtTA;shRNA-Atg5;KP. Tet-regulated shRNA expression against essential autophagy gene Atg5 that enables temporal control of Atg5 levels ubiquitously (rtTA3;shRNA-Atg5) (51) was obtained by crossing LSL-rtTA3;shRNA-Atg5 with B6.C-Tg(CMV-cre)1Cgn/J (JAX006054) mice. Subsequently, transgenic mouse line rtTA3;shRNA-Atg5 was crossed with KrasLSL-G12D/+;p53Flox/Flox GEMM to obtain systemic autophagy inducible rtTA;shRNA-Atg5;KP GEMMs for KP NSCLC. Autophagy loss switchable GEMMs for KP NSCLC are in mixture background.
KP lung tumors were induced by intranasal infection with 4x107 pfu Adenovirus-Cre per mouse as we did previously (36). Lung tumor-specific Atg5 knockdown or systemic autophagy knockdown was achieved by feeding mice with a laboratory diet containing doxycycline (Dox) at 200mg/kg (Bio-Serv Mouse Diet, S3888). Control mice were fed with a laboratory control diet (Bio-Serv Mouse Diet, S4207).
CD4 and CD8 T cell depletion
For T-cell depletion, four days before switching from normal diet to the Dox diet to systemically inhibit Atg5 expression, anti-CD4 (clone GK1.5; BE0003-1, BioXCell), anti-CD8 (clone 2.43; BE0061, BioXCell), or control IgG2b (clone LTF-2; BE0090, BioXCell) antibodies were intraperitoneally injected into KP lung tumor bearing rtTA;shRNA-Atg5, and WT-Atg5 mice, every 5 days for 3 weeks. Subsequently, mice were euthanized, and lung lobes were collected for tumor burden analysis.
[U13C6]-glucose and [U13C3]-lactate infusion
Venous catheters were surgically implanted into the jugular veins of tumor bearing mice 3-4 days prior to infusions. On the day of infusion, food was removed from the mice around 9am and infusions were started at 3pm. Infusions were performed in free-moving conscious animals. Mice were infused with [U13C6]-glucose (200mM) or [U13C3]-lactate (5%) at a rate of 0.1ul/g/min for 2.5 hours prior to euthanizing and rapid tissue harvest. Blood was collected from mouse cheek into Eppendorf™ Anticoagulant-Coated Microcentrifuge Tubes (Fisher Scientific, #05-407-13C) for plasma/serum. Lung tumors were rapidly dissected and frozen using a liquid-nitrogen cold clamp to quench metabolism and stored at −80 oC until metabolite extraction.
Histology and immunohistochemistry
Paraffin-embedded tissue sections were prepared as described previously (36) for Hematoxylin and eosin (H&E) and IHC staining. Antibodies utilized for immunohistochemistry (IHC) were CD3 (1:100 dilution, Ab16669, Abcam), CD4 (1:1000 dilution, Ab183685, Abcam), CD8 (1:100 dilution, 14-0808-82, Invitrogen), GLUT1 (1:250 dilution, Ab115730, Abcam), Atg5 (1:100 dilution, Ab108327, Abcam), p62 (1:1000 dilution, PW9860-0100, Enzo Life Sciences), p42/44 MAPK (p-ERK) (1:400 dilution, 4376, Cell Signaling), p-S6 (1:200 dilution, 4858S, Cell Signaling), Ki67 (0.1 ug/ml, Ab15580, Abcam), and cleaved caspase-3 (1:400 dilution, 9661S, Cell Signaling). For the quantification of IHC sections, tissues were analyzed by quantifying at least 10 images at 20x magnification. A minimum of 200 cells were scored for each image.
Tumor burden quantification
H&E-stained lung specimens were imaged at Rutgers Cancer Institute of New Jersey Biomedical Informatics shared resource using an Olympus VS120 whole-slide scanner (Olympus Corporation of the Americas) at 20× magnification. The image analysis protocol was custom-developed on the Visiopharm image analysis platform (Visiopharm A/S) to identify tissue area and compute tumor burden based on semi-automatically detected tumors. Tumor masks and whole-tissue masks were computed from low-resolution image maps, which were extracted from whole-slide images. Tumor and whole-tissue masks were created for each slide. The segmentation masks were used for generation of ratios of tumor burden.
Western blotting
Tissues were snap-frozen in liquid nitrogen, ground using Cryomill in liquid nitrogen at 25Hz for 2 minutes, and then lysed in Tris lysis buffer (1M Tris-HCl, 1M NaCl, 0.1M EDTA, 10% NP40). Protein concentrations were measured using the Bio-Rad BCA reagent. Samples were probed with antibodies against Atg5 (1:1000 dilution, Ab108327, Abcam), LC3 (1 ug/ml, NB600-1384, Novus Biologicals) and β-actin (1:1000 dilution, A1978, Sigma Aldrich).
Picrosirius red staining
Liver paraffin slides were de-waxed and rehydrated. Picrosirius red staining was performed as described previously (51). Slides were then dehydrated, and cover slipped for imaging.
Flow cytometry
For tumor immunoprofiling, tumors were dissected from the lung lobes of mice, and homogenized in RPMI medium (Gibco, supplemented with 10% FBS). Nonspecific binding of antibodies to cell Fc receptors was blocked using 10 μl per 107 cells of FcR blocking reagent (Miltenyi Biotec). Cell surface immunostaining was performed using the following antibodies: CD4 (1:500 dilution, clone GK1.5, 17-0041-82), CD3 (1:300 dilution, clone 17A2, 56-0032-82), CD45 (1:200 dilution, clone 30-F11, 103107) and CD8 (1:500 dilution, clone 53.67, 100749; BioLegend). The full protocol has been described previously (45). Data were acquired using an LSR-II flow cytometer (BD Biosciences) and analyzed with FlowJo software (TreeStar, San Carlos, CA).
For circulating blood immunoprofiling, peripheral blood lymphocytes (PBL) (~100 μl) was collected by retro-orbital bleeding with a calibrated micropipet coated with heparin into a Microtainer Blood Collector containing Dipotassium EDTA (Becton Dickinson, Mountain View, CA) to prevent coagulation. Red blood cells were depleted from PBL with RBC Lysis Buffer (BioLegend). Cells (1x106) were blocked with rat serum and FcR blocking Ab (2.4G2), and then incubated with various Abs conjugated to different fluorochromes for multiple color fluorescence surface staining. Cell surface immunostaining was performed with the following antibodies purchased from Biolegend: BV785-CD45, BV605-CD3, BV570-CD4, PE-Cy7-CD8a, Pacific blue-Gr-1, PE-Cy5-CD11b, APC-F4/80, and APC/Fire 750-B220. Each antibody was 1:200 dilution in Fluorescence-Activated Cell Sorting (FACS) staining buffer. List mode data were acquired on a Northern Lights spectral flow cytometer (Cytek, Fremont, CA) (52). The results were analyzed using FlowJo software (TreeStar, San Carlos, CA) (53-55)
Cytokines and chemokines assay
Cytokine and chemokine levels were determined using the Cytokine & Chemokine 26-Plex Mouse ProcartaPlex Panel 1 (EPX260-26088-901, Invitrogen). Data were collected using a Luminex-200 system (Luminex) and validated using the xPONENT software package (Luminex).
Metabolomics analysis by LC-MS
Tissue or serum metabolites extracted using methanol:acetonitrile:water (40:40:20) solution (with 0.5% formic acid solution for tissue metabolite extraction and without formic acid for serum metabolite extraction), followed by neutralization with 15% ammonium bicarbonate were used for LC-MS, as described previously (56). Samples were subjected to reversed-phase ion-pairing chromatography coupled by negative mode electrospray ionization to a stand-alone orbitrap mass spectrometer (Thermo Fisher Scientific) as described previously (56).
Statistics
Data were expressed as the mean ± s.e.m. Statistical analyses were carried out with GraphPad Prism version 9.0 or Microsoft Excel. Significance in the Kaplan-Meier analyses to determine and compare the progression-free survival was calculated using the log-rank test. The mass spectra were analyzed using MAVEN software and the peak area of each detected metabolite was obtained. Statistical significance of metabolites was determined by a paired two-tailed Student’s t-test, and >=three mice from each genotype were used. P-value of <0.05 was considered statistically significant.
Data Availability Statement
Raw data for this study were generated at Immune monitoring core facility and Metabolic core facilty at Rutgers Cancer Institute of New Jersey. Derived data supporting the findings of this study are available from the corresponding author upon request.
Results
Generation of autophagy switchable mouse models for KP NSCLC.
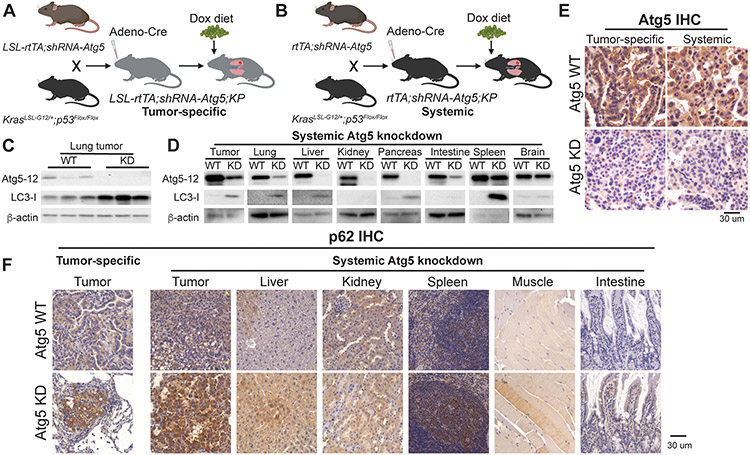
Using the KrasLSL-G12D/+;p53Flox/Flox;Atg7Flox/Flox GEMM, we demonstrated the essential role of cell autonomous autophagy to support KP lung tumorigenesis (36). However, one caveat of this model is that the deletion of Atg7 occurs simultaneously with activation of the oncogenic drivers, only happens in the tumor cells, and is irreversible, which does not fully mimic the situation of autophagy inhibition in clinical practice. To avoid above limitations, we generated autophagy-switchable mouse models for KP NSCLC through Atg5 shRNA-inducibility. Transgenic mouse lines with Tet-regulated shRNA expression against Atg5 to enable temporal control of Atg5 levels that are cell type-specific (LSL-rtTA3;shRNA-Atg5) or ubiquitous (rtTA3;shRNA-Atg5) (51) were bred with the KrasLSL-G12D/+;p53Flox/Flox GEMM to obtain the tumor-specific autophagy switchable LSL-rtTA;shRNA-Atg5;KP (Fig. 1A) and systemic autophagy switchable rtTA;shRNA-Atg5;KP (Fig. 1B) GEMMs for KP NSCLC.
Figure 1. Generation of autophagy switchable mouse models for KP NSCLC.
A and B. Scheme to generate GEMMs for KP NSCLC where Atg5 expression can specifically be modified in established lung tumors (A) or systemically in mice bearing KP lung tumors (B).
C. Western blotting for Atg5-12 and LC3-I of the lung tumors from wild type (WT) and tumor specific-Atg5 knockdown mice at three weeks post-Dox diet. β-actin serves as a protein loading control.
D. Western blotting for Atg5-12 and LC3-I of lung tumors and the indicated tissues from WT and systemic Atg5 knockdown mice at three weeks post-Dox diet. β-actin serves as a protein loading control.
E. Representative IHC for Atg5 of the lung tumors from WT mice, tumor specific-Atg5 knockdown mice and systemic Atg5 knockdown mice at three weeks post-Dox diet.
F. Representative IHC for p62 of the lung tumors from WT mice, tumor specific-Atg5 knockdown mice and systemic Atg5 knockdown mice at three weeks post-Dox diet.
For the tumor-specific switchable Atg5 knockdown, LSL-rtTA;shRNA-Atg5;KP mice were intranasally infected with Adenovirus-Cre to induce lung tumors and activate tumor-specific rtTA concurrently. At 6 weeks post-tumor induction, mice were fed with Dox diet to inhibit Atg5 expression in tumors (Fig. 1A). shRNA-Atg5;KP mice without rtTA to induce expression of Atg5 shRNA were used as the WT control. We found that Atg5-Atg12 conjugation in KP lung tumors was abolished after 3 weeks of Dox diet administration, which was accompanied by the accumulation of the autophagy substrate LC3-I (Fig. 1C), indicating autophagy blockage in tumors.
For systemic switchable Atg5 knockdown, at 6 weeks post tumor induction, rtTA;shRNA-Atg5;KP mice were fed with Dox diet to systemically inhibit Atg5 expression for 3 weeks (Fig. 1B). Col1a1-targeted shRNA transgenic mice has inefficient expression of shRNA in the brain (57). Therefore, except for brain, the reduction of Atg5-Atg12 conjugation and increased LC3-I accumulation were observed in all other tissues, including lung tumors (Fig. 1D). Most importantly, the lack of brain-associated Atg5 shRNA expression circumvents a lethal phenotype associated with complete autophagy knockouts (49,51), which provides an opportunity for longer-term analysis of autophagy loss and restoration. By IHC, we further confirmed that the extent of Atg5 knockdown in lung tumors between tumor-specific Atg5 knockdown mice and systemic Atg5 knockdown mice was comparable after 3 weeks Dox diet administration (Fig. 1E and Supp. Fig. 1). In consistent with Atg5 knockdown, p62 accumulation, an indicator of autophagy blockade, was observed in tumors and tissues in both models (Fig. 1F). In conclusion, we successfully generated novel mouse models to modulate autophagy in KP lung tumors or in system.
Systemic but not tumor-intrinsic autophagy ablation impairs established KP lung tumor growth.
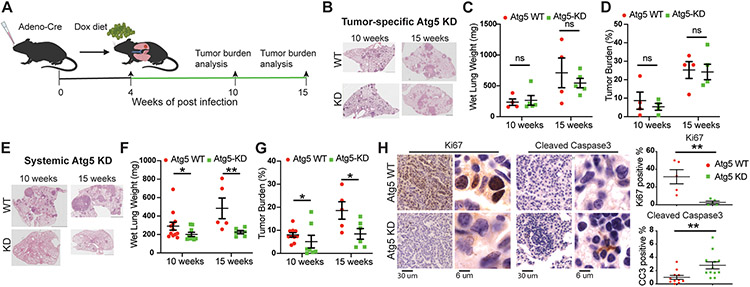
To examine the role of tumor-intrinsic and systemic autophagy in established lung tumor growth, KP lung tumors were first induced in tumor-specific Atg5 shRNA-inducible mice: LSL-rtTA;shRNA-Atg5;KP, and systemic Atg5 shRNA-inducible mice: rtTA;shRNA-Atg5;KP. At 4 weeks post-tumor induction, mice were fed with Dox diet to inhibit Atg5 expression in KP lung tumors or systemically. Subsequently, mice were sacrificed at 6 weeks and 11 weeks post-Dox treatment, and tumor burden was analyzed as done previously (Fig. 2A) (36). Compared with WT control mice, tumor-specific Atg5 knockdown did not inhibit the growth of established KP lung tumors, which was assessed by the wet lung weight and tumor burden analysis (Fig. 2B-D). Thus, the reduction of tumorigenesis by Atg7 ablation generated from the KrasLSL-G12D/+;p53Flox/Flox;Atg7Flox/Flox GEMM could be due to either more potent autophagy inhibition due to gene deletion compared to knockdown or the timing of autophagy deletion during KP lung tumorigenesis.
Figure 2. Systemic, not tumor-intrinsic, Atg5 knockdown impairs established KP lung tumor growth.
A. Scheme of the Atg5 knockdown with Dox treatment in mice bearing KP lung tumors to monitor the role of tumor cell-autonomous autophagy or systemic autophagy in established KP lung tumor growth.
B. Representative histology (H&E staining) of scanned lung sections of WT and tumor-specific Atg5 knockdown mice at the indicated time points from (A) after Dox treatment.
C. Graph of wet lung weight of WT and tumor-specific Atg5 knockdown mice at the indicated time points.
D. Quantification of tumor burden of WT and tumor-specific Atg5 knockdown mice from scanned lung sections (B).
E. Representative histology (H&E staining) of scanned lung sections of WT and systemic Atg5 knockdown mice at the indicated time points from (A) after Dox treatment.
F. Graph of wet lung weight of WT and systemic Atg5 knockdown mice at the indicated time points.
G. Quantification of tumor burden of WT and systemic Atg5 knockdown mice from scanned lung sections (E).
H. Representative IHC and quantifications of Ki67 and cleaved caspase 3 of KP lung tumors from WT and systemic Atg5 knockdown mice.
Data are mean± s.e.m. n.s., P>0.05; * P<0.05; ** P<0.01
In contrast to tumor-specific Atg5 knockdown, systemic Atg5 knockdown significantly inhibited established KP lung tumor growth. Wet lung weight and tumor burden analysis demonstrated reduced tumor growth with systemic Atg5 knockdown (Fig. 2E-G), which was accompanied by reduced cell proliferation (Ki67) and increased apoptosis (cleaved caspase-3) (Fig. 2H). These findings concur with the previously reported role for host autophagy in promoting lung tumor growth (43,45).
Acute systemic but not tumor-intrinsic autophagy inhibition impairs glucose carbon flux to major metabolic pathways of established KP lung tumors.
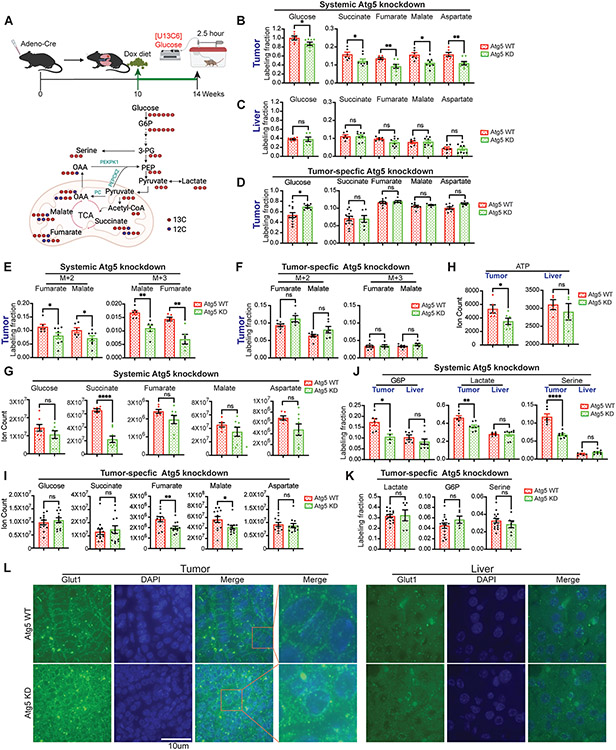
Our previous in vivo isotope tracing and metabolic flux analysis revealed that glucose is one of the major carbon sources for Kras-driven lung tumor TCA cycle metabolites (56). To gain insight into the impact of altered TCA cycle metabolism due to systemic autophagy inhibition, we performed [U13C6]-glucose in vivo tracing followed by Liquid chromatography–mass spectrometry (LC-MS) and metabolic flux analysis in mice bearing KP lung tumors with acute, tumor-specific Atg5 knockdown, or systemic Atg5 knockdown (Fig. 3A). Serum, lung tumor and liver were collected immediately after 2.5 hours of infusion for LC-MS and metabolic flux analysis as we did previously (56). Compared with Atg5 WT mice, glucose uptake in KP lung tumors was significantly lower in systemic Atg5 knockdown mice (Fig. 3B), which was accompanied by the reduced glucose carbon flux to tumor TCA cycle metabolites (Fig. 3B). However, glucose uptake and glucose carbon contribution to liver TCA intermediates were not altered by systemic Atg5 knockdown (Fig. 3C). In contrast, acute, tumor-specific Atg5 knockdown had no effect on glucose carbon flux to tumor TCA cycle intermediates (Fig. 3D). These findings suggest that systemic Atg5 knockdown, not tumor-intrinsic Atg5 knockdown, specifically affects the glucose metabolism in KP lung tumors.
Figure 3. Acute systemic, not tumor-intrinsic, autophagy inhibition impairs glucose carbon flux to major metabolic pathways of established KP lung tumors.
A. Scheme of the 2.5 hour in vivo [U13C6]-glucose tracing in KP lung tumor bearing mice (top panel) and carbon contribution from glucose to glycolytic intermediates, TCA cycle intermediates and serine (bottom panel).
B. Normalized labelling fraction of glucose and TCA cycle intermediates of KP lung tumors in WT and systemic Atg5 knockdown mice in fasted state.
C. Normalized labelling fraction of glucose and TCA cycle intermediates of liver in WT and systemic Atg5 knockdown mice in fasted state.
D. Normalized labelling fraction of glucose and TCA cycle intermediates of KP lung tumors in WT and tumor-specific Atg5 knockdown mice in fasted state.
E. Normalized labelling fraction of M+2 and M+3 fumarate and malate of lung tumors in WT and systemic Atg5 knockdown mice in fasted state.
F. Normalized labelling fraction of M+2 and M+3 fumarate and malate of lung tumors in WT and tumor-specific Atg5 knockdown mice in fasted state.
G. Levels of glucose and TCA cycle metabolites in KP lung tumors of WT and systemic Atg5 knockdown mice after 4 weeks Dox treatment in fasted state.
H. Levels of ATP in KP lung tumors of WT and systemic Atg5 knockdown mice after 4 weeks Dox treatment in fasted state.
I. Levels of glucose and TCA cycle metabolites in KP lung tumors of WT and tumor-specific Atg5 knockdown mice after 4 weeks Dox treatment in fasted state.
J. Normalized labelling fraction of G6P, lactate, and serine in KP lung tumors and liver of WT and systemic Atg5 knockdown mice in fasted state.
K. Normalized labelling fraction of G6P, lactate, and serine in KP lung tumors of WT and tumor-specific Atg5 knockdown mice in fasted state.
L. Immunofluorescence of Glut1 of KP lung tumors and liver from WT and systemic Atg5 knockdown mice after 4 weeks Dox treatment (Green: Glut1; Blue: DAPI for nucleus).
Labeling fraction are normalized to circulating serum [U13C6]-glucose. Data are mean± s.e.m. n.s., P>0.05; * P<0.05; ** P<0.01; *** P<0.005; **** P<0.001
Glucose can contribute carbon to TCA cycle intermediates via converting glucose-derived pyruvate to acetyl-CoA mediated by the pyruvate dehydrogenase complex, resulting in isotopomer species with two labeled carbons (M+2) (Fig. 3A). TCA cycle anaplerosis involving glucose-derived carbon and pyruvate carboxylase has been reported in some human lung tumors (58,59), leading to the increased three carbon labeling (M+3) of TCA cycle metabolites from glucose (Fig. 3A) (60). We found that systemic Atg5 knockdown significantly reduced the fraction of both M+2 and M+3 carbon labeling from glucose to TCA cycle metabolites of lung tumors (Fig. 3E). However, when Atg5 was specifically knocked down in lung tumors, both M+2 and M+3 carbon labelings from glucose to tumor TCA cycle metabolites were not affected (Fig. 3F). Consistent with the reduced glucose carbon flux to tumor TCA cycle intermediates, there was a trend of decreased level of most TCA cycle metabolites in KP lung tumors due to acute systemic Atg5 knockdown (Fig. 3G), leading to significantly lower ATP level in lung tumors compared with WT mice (Fig. 3H). We observed that the levels of fumarate and malate were decreased by tumor-specific Atg5 knockdown (Fig. 3I), although tumor-specific Atg5 knockdown did not alter glucose flux to TCA cycle intermdiates (Fig. 3D & F).
Besides supporting mitochondrial TCA cycle metabolism, glucose contributes carbons for biosynthesis via incorporation of glycolytic intermediates to different metabolic pathways (Fig. 3A). Acute systemic Atg5 knockdown significantly decreased the carbon contribution to glycolytic intermediates, for example, glucose 6-phosphate (G6P) and lactate. Moreover, labeling of serine in KP lung tumors was significantly lower in Atg5 knockdown mice than that in WT mice (Fig. 3J). However, such alteration was not observed in the liver from mice with systemic Atg5 knockdown compared with WT mice (Fig. 3K). Moreover, tumor-specific Atg5 knockdown had no impact on glucose carbon contribution to lactate, G6P and serine (Fig. 3K).
In response to metabolic stresses and RAS activation, autophagy upregulation not only drives intracellular degradation (32), but also facilitates glucose uptake and glycolytic flux by promoting cell surface trafficking of a key nutrient transporter, GLUT1 in MEF cells (61). We found that cell surface expression of Glut1 was disrupted in KP lung tumors in mice with acute systemic Atg5 knockdown compared with tumors in WT mice. Conversely, more Glut1 puncta were observed in the lung tumor cytoplasm of acute systemic Atg5 knockdown mice (Fig. 3L). Thus, decreased glucose uptake in KP lung tumors of systemic Atg5 knockdown mice may be due to lack of membrane Glut1 trafficking. It has been reported that GLUT1 membrane localization is restricted to hepatocytes proximal to the hepatic venule; Instead, GLUT2 fulfils the major glucose transport role in hepatocytes (62). As expected, the Glut1 expression was not detected in liver by immunofluorescence in both autophagy intact and systemic autophagy knockdown mice (Fig. 3L).
Systemic but not tumor-intrinsic autophagy ablation inhibits KP lung tumor gluconeogenesis and biosynthesis.
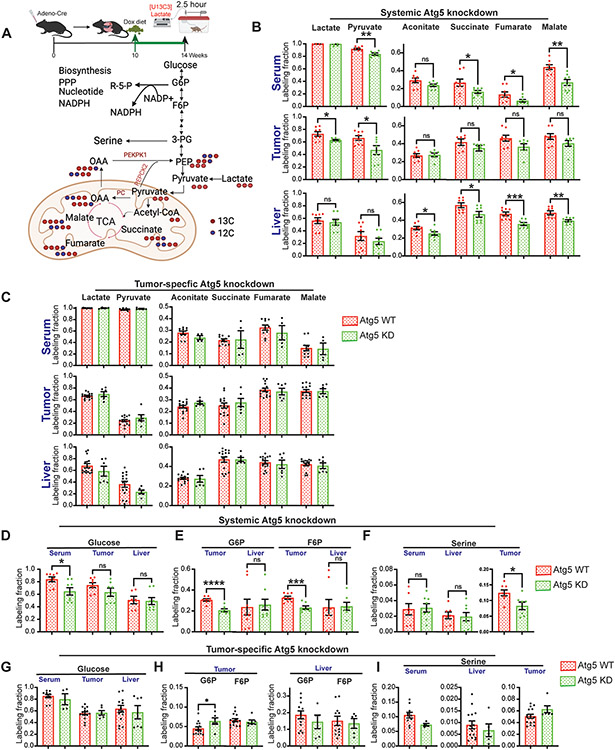
We have previously demonstrated that glucose feeds KP lung tumor TCA cycle metabolites via circulating lactate (56). We therefore performed [U13C3]-lactate in vivo tracing followed by LC-MS and metabolic flux analysis in mice bearing KP lung tumors (Fig. 4A) to examine the direct carbon source from lactate to tissue TCA cycle intermediates caused by systemic Atg5 knockdown or tumor-specific Atg5 knockdown. Serum, lung tumor and liver were collected for LC-MS and metabolic flux analysis. Despite significantly reduced lactate uptake and carbon flux from lactate to pyruvate in KP lung tumors with systemic Atg5 knockdown (Fig. 4B), the direct carbon contribution from lactate to tumor TCA cycle metabolites was only slightly reduced (Fig. 4B, Supp. Fig. 2). However, the labeled TCA cycle intermediates from lactate in circulating serum and liver were significantly lower in systemic Atg5 knockdown mice compared with WT mice (Fig. 4B, Supp. Fig. 2), which suggests that autophagy is essential to maintain liver TCA cycle metabolism. Moreover, Atg5 knockdown in tumor alone had no effect on lactate carbon flux to TCA cycle intermediates of serum, lung tumor and liver (Fig. 4C). This further supports the concept that the effect of systemic autophagy inhibition on the metabolism is tumor- and tissue-specific.
Figure 4. Systemic autophagy ablation, not tumor-intrinsic, autophagy inhibition impairs tumor gluconeogenesis and biosynthesis.
A. Scheme of the 2.5 hour in vivo [U13C3]-lactate tracing in KP lung tumor bearing mice (top panel) and carbon contribution from lactate to TCA cycle intermediates, glycolytic intermediates, and serine (bottom panel).
B. Normalized labelling fraction of lactate, pyruvate and TCA cycle intermediates of serum, lung tumor and liver in WT and systemic Atg5 knockdown mice in fasted state.
C. Normalized labelling fraction of lactate, pyruvate and TCA cycle intermediates of serum, lung tumor and liver in WT and tumor-specific Atg5 knockdown mice in fasted state.
D. Normalized labelling fraction of glucose of serum, lung tumors and liver in WT and systemic Atg5 knockdown mice in fasted state.
E. Normalized labelling fraction of G6P and F6P of lung tumors and liver in WT and systemic Atg5 knockdown mice in fasted state.
F. Normalized labelling fraction of serine of serum, liver and lung tumors in WT and systemic Atg5 knockdown mice in fasted state.
G. Normalized labelling fraction of glucose of serum, lung tumors and liver in WT and tumor-specific Atg5 knockdown mice in fasted state.
H. Normalized labelling fraction of G6P and F6P of lung tumors and liver in WT and tumor-specific Atg5 knockdown mice in fasted state.
I. Normalized labelling fraction of serine of serum, liver and lung tumors in WT and tumor-specific Atg5 knockdown mice in fasted state.
Labeling fraction are normalized to circulating serum [U13C3]-lactate. Data are mean± s.e.m. n.s., P>0.05; * P<0.05; ** P<0.01; *** P<0.005; **** P<0.001
Gluconeogenesis, at least a partial pathway from the TCA cycle to glycolytic intermediates, occurs throughout the body (63). Cancer has been reported to hijack key gluconeogenic enzymes, phosphoenolpyruvate carboxykinase (PEPCK), fructose-1,6-bisphosphatase, and glucose-6-phosphatase, to engage in truncated gluconeogenesis, which promotes metabolic flexibility by allowing the utilization of non-carbohydrate sources for biosynthesis and redistributing glucose flux for antioxidant production (64-66) (Fig. 4A). Acute, systemic Atg5 knockdown had no impact on glucose labeling from lactate in tumor and liver (Fig. 4D). However, the labeling fraction of glycolytic intermediates G6P and fructose 6-phosphate (F6P) in KP lung tumors from systemic Atg5 knockdown mice was significantly lower than that in WT mice; while there was no difference in the G6P and F6P labeling fraction of the liver between systemic Atg5 knockdown mice and WT mice (Fig. 4E). Moreover, the labeling fraction of tumor serine, not liver serine, from lactate was significantly reduced by systemic Atg5 knockdown (Fig. 4F). This phenomenon was not observed when Atg5 was specifically knocked down in established KP lung tumors (Fig. 4G-I). Lactate is one of the gluconeogenic precursors (63). Our in vivo tracing study suggests that systemic autophagy could support KP lung tumor growth by hijacking truncated gluconeogenesis for biosynthesis of serine.
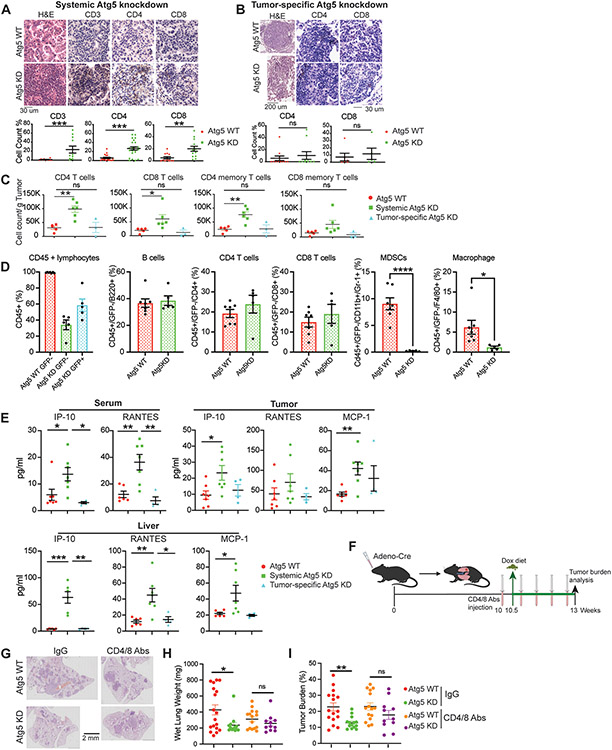
Systemic autophagy suppresses tumor killing by T cells in established KP lung tumors.
Patients with NSCLC show better prognosis when tumors are associated with higher lymphocyte infiltration (67,68). Compared with control mice, more lymphocyte infiltration was observed in KP lung tumors of mice with acute, systemic Atg5 knockdown for four weeks as revealed by H&E staining and IHC for CD3, CD4 and CD8 T cells (Fig. 5A). However, tumor-specific Atg5 knockdown had no impact on tumor T cell infiltration (Fig. 5B). We further validated immune profiling of the KP lung tumor microenvironment via multiplex immunofluorescent staining and flow cytometry. The percentages of tumor infiltrating CD4, CD8 T cells and CD4 memory T cells were significantly higher in tumors upon systemic Atg5 knockdown than with tumor cell-specific Atg5 knockdown or WT mice after 3 weeks of Dox treatment (Fig. 5C). We next examined the circulating lymphocyte profiling via multiplex immunofluorescent staining and flow cytometry. The feature of systemic autophagy inducible mouse model is that Atg5shRNA expression can be detected by the GFP. Interestingly, we found that at four weeks post-Dox treatment, about 40% of CD45+ lymphocytes showed GFP negative (GFP−), indicating autophagy intact CD45+ lymphocytes. We next compared those CD45+/GFP− lymphocytes in circulation between WT control and systemic Atg5 knockdown mice. We found that there were no significant differences in B cells, CD4 T cells and CD8 T cells in the circulation between control and systemic Atg5 knockdown mice; however, the percentages of myeloid-derived suppressor cells (MDSCs) and macrophages were significantly lower in the circulation of systemic Atg5 knockdown mice than control mice (Fig. 5D).
Figure 5. Systemic autophagy suppresses T cell-mediated tumor killing for KP lung tumor growth.
A. Representative H&E staining, IHC images and quantifications of CD3, CD4 and CD8 T cells in lung tumors of WT and systemic Atg5 knockdown mice.
B. Representative H&E staining, IHC images and quantifications of CD4 and CD8 T cells in lung tumors of WT and tumor-specific Atg5 knockdown mice.
C. CD4, CD8, CD4 memory and CD8 memory T cells in KP lung tumors of WT, systemic Atg5 knockdown, and tumor-specific Atg5 knockdown mice examined via multiplex immunofluorescent staining and flow cytometry sorting at four weeks post Dox treatment.
D. Circulating blood immunoprofiling of WT and systemic Atg5 knockdown mice examined via multiplex immunofluorescent staining and flow cytometry sorting at four weeks post Dox treatment.
E. Serum, tumor and liver cytokine and chemokine profiling of WT, systemic Atg5 knockdown, and tumor-specific Atg5 knockdown mice at four weeks post Dox treatment.
F. Scheme of co-depletion of CD4 and CD8 T cells to assess the role of systemic autophagy in modulating immune response for established KP lung tumor growth.
G. Representative histology (H&E staining) of scanned lung sections of WT and systemic Atg5 knockdown mice without or with CD4/CD8 T cell depletion.
H. Graph of wet lung weight of WT and systemic Atg5 knockdown mice without or with CD4/CD8 T cell depletion.
I. Quantification of tumor burden of WT and systemic Atg5 knockdown mice without or with CD4/CD8 T cell depletion from (F).
Data are mean± s.e.m. n.s., P>0.05; * P<0.05; ** P<0.01; *** P<0.005
Host autophagy has been reported to support allograft tumor growth via autophagy-mediated “hepatic autophagy immune tolerance”, thereby inhibiting anti-tumor T-cell responses (45). We therefore performed multiplex cytokine analysis in serum, liver and tumor in KP lung tumor bearing mice after three weeks of Dox treatment. Consistent with our previous findings (45,49), we found that IFN-γ-inducible protein 10 (IP-10/CXCL10) and CCL5 (regulated upon activation, normal T cell expressed and secreted, RANTES) were significantly increased in serum, tumor and liver in KP lung tumor bearing mice with systemic Atg5 knockdown compared with WT mice and tumor-specific Atg5 knockdown mice. Additionally, we found that the level of monocyte chemoattractant protein-1 (MCP-1/CCL2) was significantly higher in liver and lung tumors with systemic Atg5 knockdown than that in WT mice (Fig. 5E).
The alteration in the immune profile of the tumor microenvironment and changes of systemic cytokines/chemokines prompted us to examine the role of systemic autophagy-mediated immune response in KP lung tumor growth by co-immunodepletion of CD4 and CD8 T cells. We first induced KP lung tumors in systemic Atg5 shRNA-inducible mice: rtTA;shRNA-Atg5;KP. At 10 weeks post-tumor induction, mice were treated with Dox to systemically inhibit Atg5 expression; together with CD4 and CD8 T cells co-immunodepletion with anti-CD4 and CD8 antibodies (200ug/mouse, i.p. every 5 days) (Fig. 5F). After 3 weeks of treatment, as expected, systemic Atg5 knockdown significantly reduced KP lung tumor growth compared with control mice when mice were treated with control IgG, which was abolished by the co-depletion of CD4 and CD8 T cells (Fig. 5G-I), indicating that one of the mechanisms of systemic autophagy-mediated KP lung tumor growth is through promoting immune evasion.
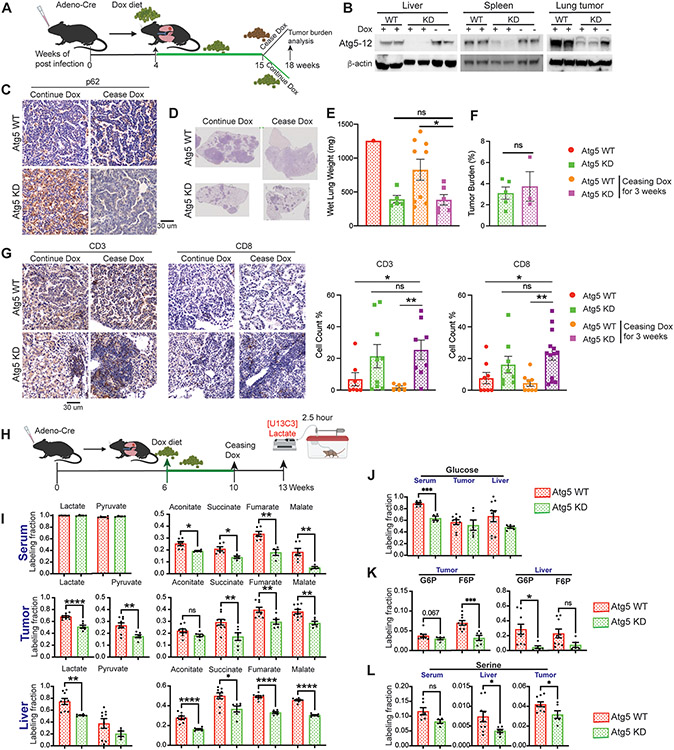
Inhibition of tumorigenesis with short-term systemic autophagy ablation is irreversible.
To evaluate the reversibility of loss of Atg5, we assessed if Atg5 knockdown and then restoration allowed for recovery of KP lung tumor growth. At 15 weeks post-Dox treatment, we ceased Dox for 3 weeks to restore Atg5 (Fig. 6A). We observed that Atg5 expression was restored in most tissues, including KP lung tumors (Fig. 6B). The accumulation of p62 in lung tumors also disappeared after ceasing the Dox diet (Fig. 6C). Excitingly, inhibition of KP tumor growth caused by knockdown of Atg5 was not rescued upon restoration of Atg5 expression for three weeks (Fig. 6D-F). Moreover, the increased CD3 and CD8 T cells in KP lung tumor microenvironment were maintained when systemic autophagy was restored in the short term (Fig. 6G).
Figure 6. Transient systemic autophagy ablation irreversibly inhibits KP lung tumor growth.
A. Scheme of the Atg5 knockdown with Dox treatment and Atg5 restoration by ceasing Dox to monitor KP lung tumor growth in systemic Atg5 shRNA inducible mice.
B. Western blotting for Atg5-12 of liver, spleen and tumor from WT and systemic Atg5 shRNA inducible mice to indicate Atg5 re-expression after ceasing Dox treatment. β-actin serves as a protein loading control.
C. Representative IHC for p62 of the lung tumors from WT mice and systemic Atg5 shRNA inducible mice.
D. Representative histology (H&E staining) of scanned lung sections of WT and systemic Atg5 shRNA inducible mice after ceasing Dox treatment.
E. Graph of wet lung weight of WT and systemic Atg5 shRNA inducible mice after ceasing Dox treatment.
F. Quantification of tumor burden of WT and systemic Atg5 shRNA inducible mice after ceasing Dox treatment.
G. Representative IHC images and quantifications of CD3 and CD8 T cells in lung tumors of WT and systemic Atg5 shRNA inducible mice.
H. Scheme of the 2.5 hour in vivo [U13C3]-lactate tracing in KP lung tumor bearing mice.
I. Normalized labelling fraction of lactate, pyruvate and TCA cycle intermediates of serum, lung tumor and liver in WT and systemic Atg5 knockdown mice after Atg5 restoration.
J. Normalized labelling fraction of glucose of serum, lung tumors and liver in WT and systemic Atg5 knockdown mice after Atg5 restoration.
K. Normalized labelling fraction of G6P and F6P of serum, liver and lung tumors in WT and systemic Atg5 knockdown mice after Atg5 restoration.
L. Normalized labelling fraction of serine of serum, liver and lung tumors in WT and systemic Atg5 knockdown mice after Atg5 restoration.
Labeling fraction are normalized to circulating serum [U13C3]-lactate. Data are mean± s.e.m. * P<0.05; ** P<0.01; *** P<0.005; **** P<0.001
To determine whether altered tumor metabolism can be sustained after Atg5 was re-expressed, we performed [U13C3]-lactate in vivo tracing and metabolic flux analysis after 4 weeks Dox diet followed by ceasing Dox diet for three weeks (Fig. 6H). In addition to maintaining a reduced carbon flux from lactate to serum and liver TCA cycle metabolites, we observed that the contribution of lactate carbon to the tumor TCA cycle intermediates was attenuated after short-term autophagy recovery (Fig. 6I). Moreover, the reduced labeling fraction of tumor G6P, F6P and serine from lactate was sustained after short-term autophagy restoration (Fig. 6J-L). Thus, altered tumor gluconeogenesis and biosynthesis by acute systemic autophagy ablation was maintained once Atg5 expression is restored in the short term.
Taken together, the increased CD8 T cell infiltration in KP lung tumor microenvironment and reduced tumor gluconeogenesis and biosynthesis due to systemic Atg5 knockdown were sustained when systemic autophagy was restored in the short term. This may contribute to irreversible tumor regression in the short term of systemic autophagy restoration.
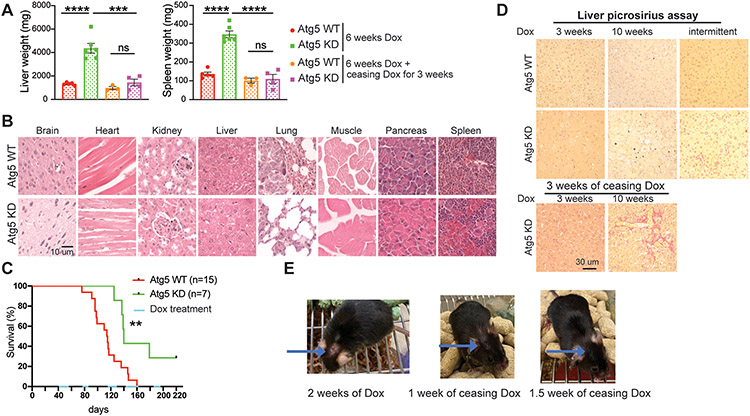
Damage of normal tissues caused by short-term systemic autophagy inhibition is reversible.
Autophagy is essential for normal tissue homeostasis with some tissues such as liver having a greater dependence on autophagy than others (23,49,69). Compared to normal tissues, the preferential sensitivity of tumors to autophagy inhibition is a prerequisite for using autophagy inhibitors as a cancer treatment. Therefore, we also examined the histology of normal tissues with systemic loss of autophagy due to Atg5 shRNA expression and restoration in mice with KP lung tumors. As expected (49,51), liver and spleen weight were increased with Atg5 knockdown in mice at 6 weeks post Dox treatment, and these levels returned to normal when autophagy was restored by ceasing Dox diet for 3 weeks (Fig. 7A). Irreversible Atg7 knockout in the host results in the release of ARG1 from the liver and the degradation of circulating arginine, which is essential for tumor growth (43). However, serum arginine levels were not significantly different between Atg5 knockdown mice and WT mice (Supp. Fig. 3A), indicating less damage to the liver with short term Atg5 knockdown compared to irreversible autophagy gene deletion. As a result, arginine level in KP lung tumors of Atg5 knockdown mice was the same as in WT mice (Supp. Fig. 3B). We did not observe damage to other tissues when systemic autophagy was inhibited with Atg5 knockdown within 6 weeks (Fig. 7B).
Figure 7. Damage induced by short-term systemic autophagy ablation is reversible in normal tissues.
A. Graphs of wet liver weight and wet spleen weight of WT and systemic Atg5 shRNA inducible mice without or with cessation of Dox treatment.
B. Representative H&E staining of indicated tissues from WT and Atg5 shRNA inducible mice after 3 weeks Dox treatment.
C. Kaplan-Meier survival curve of WT and systemic Atg5 shRNA inducible mice that were intranasally infected with Adenovirus-Cre, followed by intermittent Dox treatment.
D. Representative images of picrosirius red staining in liver sections of WT and systemic Atg5 shRNA inducible mice with continued 3 weeks, 10 weeks Dox treatment, then followed by cessation of Dox for 3 weeks, and intermittent Dox treatments as indicated in (C).
E. Representative pictures of systemic Atg5 knockdown lung tumor bearing mice showing that cessation of Dox treatment abolished skin side effects caused by systemic Atg5 knockdown.
Data are mean± s.e.m. n.s., P>0.05; * P<0.05; ** P<0.01; *** P<0.005; **** P<0.001
To avoid the normal tissue damage from long term autophagy ablation, when KP lung tumors were established, we modulated systemic autophagy status (on-off-on) by intermittent Dox diet administration and assessed mouse survival. Compared with control mice, transient systemic Atg5 knockdown significantly extended the mouse life span (Fig. 7C). Liver fibrosis was observed with restoration of autophagy after long-term systemic Atg5 knockdown (50). We therefore examined liver fibrosis using picrosirius red staining. We as well observed liver fibrosis in mice after restoration of Atg5 following long (continuous 10 weeks) autophagy ablation (Fig. 7D). However, we did not observe liver fibrosis when the systemic Atg5 was intermittently inhibited in the short-term (3 weeks) (Fig. 7D).
Whole body-Atg5 knockdown mice are known to be susceptible to eye infection and ulcerative dermatitis (51). Such phenotype was observed in about 5% of KP lung tumor bearing mice when systemic Atg5 was knocked down, which completely healed after ceasing Dox for 10 days to restore autophagy (Fig. 7E). Thus, comparing and contrasting the consequences of autophagy on-off-on modulation in tumor versus normal tissues from the same mouse (Fig. 6 & Fig. 7), we conclude that KP lung tumors are more sensitive to acute autophagy inhibition than normal tissues, which provides a therapeutic window for inhibiting autophagy to treat KP lung tumors.
Discussion
Autophagy-mediated recycling of cellular contents in lysosomes provides necessary metabolites for pivotal metabolic pathways and suppresses inflammation through clearance of damaged proteins and organelles. Cancer cells hijack autophagy to maintain metabolism, growth survival and malignancy and to evade an anti-tumor T-cell response (21). Using Cre-Flox mouse models for Kras-driven NSCLC, we have demonstrated that autophagy is required for KP lung tumorigenesis (36,49). However, deletion of Atg7 that occurs at the same time as KP lung tumor initiation prevents us from elucidating the role of tumor cell autonomous autophagy in the growth of established KP lung tumors. In addition, irreversible deletion of essential autophagy genes in adult mice causes damage to some normal tissues, which limits our further evaluation of the reversibility of that damage caused by systemic autophagy ablation. Whether to translate autophagy inhibition for Kras-driven lung cancer therapy requires an understanding of the differential sensitivity of tumor versus normal tissue to autophagy inhibition and the potential differential reversibility of that sensitivity. Therefore, we took advantage of recently developed autophagy inhibition-inducible mouse models, in which autophagy can be dynamically modulated tissue-specifically or ubiquitously via inducible expression of shRNA targeting Atg5 (51), to study the role of tumor-cell autonomous and systemic autophagy in the growth of established KP lung tumors. Most importantly, the lack of brain-associated shRNA expression in this model circumvents the lethal phenotype associated with complete autophagy knockouts, which allows us to understand the potential adverse effects of switching systemically from an autophagy suppressed and restored state during cancer treatment.
In contrast to deletion of essential autophagy gene in tumor cells at the point of tumor initiation, which inhibits tumor growth (36), we found that acute tumor-specific Atg5 knockdown does not inhibit the growth of established KP lung tumors. This may be explained by less robust loss of autophagy function with knockdown compared to gene deletion or by the different timing of autophagy ablation, which requires further investigation. Moreover, we found that systemic Atg5 knockdown significantly inhibits KP lung tumor growth and extends mouse life span, which is in consistent with our previous findings reporting a critical role of host autophagy in promoting tumor growth (49).
In contrast to Cre-Flox GEMMs in which essential autophagy genes are deleted irreversibly (36,49), a mouse model of systemic autophagy inhibition-inducible with conditional Atg5 shRNA expression allows us to examine the reversibility of autophagy deficiency in normal and tumor tissue. It also enables a direct comparison of acute autophagy depletion and its reversibility in both normal tissues and tumor simultaneously. Importantly, we found that inducible systemic suppression of Atg5 was more toxic to lung tumors than to normal tissues, and that normal tissues but not lung tumors recovered from that damage upon restoration of autophagy function in a short term. However, although toggling Atg5 on-off-on by intermittently treating mice with Dox significantly extended mouse life span, the mouse still died from lung tumors, suggesting that KP lung tumor might continue to grow after long term autophagy restoration. Therefore, autophagy inhibition combined with other therapies, such as immunotherapy or targeted therapy, could be a better option for KP lung cancer treatment. Thus, NSCLC is selectively autophagy dependent, and in contrast to normal tissues, the resulting damage to tumors is irreversible when autophagy is restored for a short term. Most importantly, the transient autophagy deficiency and restoration described for the first time here resembles what would occur with autophagy inhibition for cancer therapy. The pinnacle observation was that toggling Atg5 on-off-on as what would occur in cancer treatment with an autophagy inhibitor substantially impaired tumorigenesis and increased mouse lifespan with limited damage to normal tissues indicative of a therapeutic window.
Glucose and lactate are two major carbon sources for Kras-driven lung tumor TCA cycle metabolites (56,70). Via in vivo 13C isotope tracing and metabolic flux analyses in both tumor-specific autophagy inducible mouse model and systemic autophagy inducible mouse model, we found that transient systemic autophagy ablation, not tumor-specific autophagy inhibition, selectively reduced the glucose and lactate uptake specifically in tumors, resulting in a decreased glucose carbon flux to tumor TCA cycle intermediates and less tumor ATP production. Cancer cells hijack gluconeogenesis for biosynthesis and redistributing glucose flux for antioxidant production and biosynthesis (64-66). Truncated gluconeogenesis is observed in KP lung tumors, as evidenced by 13C carbon labeling from [U13C3]-lactate to G6P, F6P and serine, which is impaired by transient systemic Atg5 knockdown. Another explanation could be that tumor cells upregulate the uptake of circulating glycolytic intermediates for biosynthesis, which can be dampened by systemic Atg5 knockdown. Indeed, systemic autophagy inhibition selectively inhibits lactate and glucose uptake in KP lung tumors, but not in liver. In response to metabolic stresses and RAS activation, autophagy facilitates glucose uptake by promoting cell surface trafficking of GLUT1 in MEF cells (61). We found that cell surface expression of Glut1 was disrupted in KP lung tumors in mice with acute systemic Atg5 knockdown, which might partially affect glucose uptake in KP lung tumors. Besides Glut1, systemic autophagy inhibition could prevent nutrient uptake by modulating other nutrient transporters in KP lung tumors, such as MCT1/4 (lactate transporter), which requires further investigation. Most importantly, altered tumor metabolism by acute systemic autophagy ablation was maintained when Atg5 expression was restored for a short term.
In addition to promoting tumor metabolism, autophagy enables cancer cells to escape immune surveillance in several ways. Autophagy downregulates cell surface expression of MHC-I limiting antigen presentation and an anti-tumor T-cell response in PDAC (35). Host autophagy maintains liver integrity to prevent type I and II interferon-mediated T-cell killing of allografted tumors (45). Inflammatory chemokines are indispensable “gate keepers” of immunity and inflammation. Consistent with our previous findings (45), systemic Atg5 knockdown in KP tumor-bearing mice led to the changes in systemic cytokines/chemokines, including increased IP-10 and CCL5 in serum and liver and increased MCP-1 in in both liver and KP lung tumors. IP-10, secreted in response to IFN-γ, plays a role in effector T Cell generation and trafficking (71), promotes damage in established tumor vasculature and inhibits human NSCLC tumorigenesis and spontaneous metastases (72,73). High serum CCL5 level is associated with better disease-free survival of patients with early breast cancer (74). MCP-1 contributes to the recruitment of blood monocytes into sites of inflammatory responses and tumors (75). In circulation, the percentage of autophagy intact CD45+ lymphocytes, including B cells, CD4 T cells and CD8 T cells are similar between control and systemic Atg5 knockdown mice, which could be due to insufficient Atg5 knockdown or generation of new lymphocytes to compensate for the loss of functional CD4/8/B cells due to autophagy ablation. Furthermore, depletion of CD4/8 T cells partially rescued defective KP lung tumor growth caused by systemic Atg5 knockdown, demonstrating that one of mechanisms by which systemic autophagy promotes KP lung tumor growth is through preventing tumor killing by T cells. As normal tissues recover from damage induced by transient loss and restoration of autophagy whereas tumors do not, perhaps it is the persistent damage in tumors that sustains an anti-tumor immune response.
Supplementary Material
Statement of Significance.
Transient loss of systemic autophagy causes irreversible damage to tumors by suppressing cancer cell metabolism and promoting anti-tumor immunity, supporting autophagy inhibition as a rational strategy for treating lung cancer.
Acknowledgments
We are grateful to Xuefei Luo, Amy Lee, Nuha Syed, Akash Raju, Jerry Kong and Michael Rangel in the Guo laboratory for their assistance with mouse ear tagging and genotyping. This work was supported by National Institute of Health (NIH) grant R01CA237347, K22CA190521, American Cancer Society grant 134036-RSG-19-165-01-TBG, GO2 Foundation for Lung Cancer, and Ludwig Princeton Branch of the Ludwig Institute for Cancer Research to J Guo; NIH grant R01CA163591 and Ludwig Princeton Branch of the Ludwig Institute for Cancer Research to E White; New Jersey Commission on Cancer Research (NJCCR) grant DFHS18PPC021 to K Khayati; NJCCR grant DCHS19PPC013, a scholarship from the Cox Foundation for Cancer Research and Mistletoe Research Fellowship to V Bhatt; NJCCR grant COCR22PDF009 to F Alogaili; NIH P30 CA072720 to Rutgers Cancer Institute of New Jersey; Cancer Research UK Cambridge Institute core grant C9545/A29580 and Biotechnology and Biological Sciences Research Council (BBSRC) BB/S013466/1 to M Narita; BBSRC BB/T013486/1 to L Cassidy and M Narita.
Footnotes
Potential Conflicts of Interest
E. W. is a stockholder in Forma Therapeutics and a founder of Vescor Therapeutics.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5–29 [DOI] [PubMed] [Google Scholar]
- 2.Barbosa R, Acevedo LA, Marmorstein R. The MEK/ERK Network as a Therapeutic Target in Human Cancer. Mol Cancer Res 2021;19:361–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han J, Liu Y, Yang S, Wu X, Li H, Wang Q. MEK inhibitors for the treatment of non-small cell lung cancer. J Hematol Oncol 2021;14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim C, Giaccone G. MEK inhibitors under development for treatment of non-small-cell lung cancer. Expert Opin Investig Drugs 2018;27:17–30 [DOI] [PubMed] [Google Scholar]
- 5.Patnaik A, Kang SP, Rasco D, Papadopoulos KP, Elassaiss-Schaap J, Beeram M, et al. Phase I Study of Pembrolizumab (MK-3475; Anti-PD-1 Monoclonal Antibody) in Patients with Advanced Solid Tumors. Clinical cancer research : an official journal of the American Association for Cancer Research 2015;21:4286–93 [DOI] [PubMed] [Google Scholar]
- 6.Robert C, Ribas A, Wolchok JD, Hodi FS, Hamid O, Kefford R, et al. Anti-programmed-death-receptor-1 treatment with pembrolizumab in ipilimumab-refractory advanced melanoma: a randomised dose-comparison cohort of a phase 1 trial. Lancet 2014;384:1109–17 [DOI] [PubMed] [Google Scholar]
- 7.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018–28 [DOI] [PubMed] [Google Scholar]
- 8.Dong ZY, Zhong WZ, Zhang XC, Su J, Xie Z, Liu SY, et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research 2017;23:3012–24 [DOI] [PubMed] [Google Scholar]
- 9.Cortez MA, Ivan C, Valdecanas D, Wang X, Peltier HJ, Ye Y, et al. PDL1 Regulation by p53 via miR-34. J Natl Cancer Inst 2016;108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji M, Liu Y, Li Q, Li X, Ning Z, Zhao W, et al. PD-1/PD-L1 expression in non-small-cell lung cancer and its correlation with EGFR/KRAS mutations. Cancer Biol Ther 2016;17:407–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uras IZ, Moll HP, Casanova E. Targeting KRAS Mutant Non-Small-Cell Lung Cancer: Past, Present and Future. Int J Mol Sci 2020;21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRAS(G12C) Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer discovery 2020;10:54–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skoulidis F, Li BT, Dy GK, Price TJ, Falchook GS, Wolf J, et al. Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N Engl J Med 2021;384:2371–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019;575:217–23 [DOI] [PubMed] [Google Scholar]
- 15.Kim D, Xue JY, Lito P. Targeting KRAS(G12C): From Inhibitory Mechanism to Modulation of Antitumor Effects in Patients. Cell 2020;183:850–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukhopadhyay S, Vander Heiden MG, McCormick F. The Metabolic Landscape of RAS-Driven Cancers from biology to therapy. Nat Cancer 2021;2:271–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017;168:657–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci 2010;35:427–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White E, Mehnert JM, Chan CS. Autophagy, Metabolism, and Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2015;21:5037–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov 2011;10:671–84 [DOI] [PubMed] [Google Scholar]
- 21.White E, Lattime EC, Guo JY. Autophagy Regulates Stress Responses, Metabolism, and Anticancer Immunity. Trends Cancer 2021;7:778–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rangel M, Kong J, Bhatt V, Khayati K, Guo JY. Autophagy and tumorigenesis. FEBS J 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol 2010;12:823–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collier JJ, Guissart C, Olahova M, Sasorith S, Piron-Prunier F, Suomi F, et al. Developmental Consequences of Defective ATG7-Mediated Autophagy in Humans. N Engl J Med 2021;384:2406–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White E, Karp C, Strohecker AM, Guo Y, Mathew R. Role of autophagy in suppression of inflammation and cancer. Current opinion in cell biology 2010;22:212–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider JL, Cuervo AM. Autophagy and human disease: emerging themes. Curr Opin Genet Dev 2014;26:16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang P, Mizushima N. Autophagy and human diseases. Cell Res 2014;24:69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006;441:880–4 [DOI] [PubMed] [Google Scholar]
- 29.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes & development 2007;21:1367–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009;137:1062–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes & development 2011;25:795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Molecular biology of the cell 2011;22:165–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes & development 2011;25:460–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes & development 2011;25:717–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto K, Venida A, Yano J, Biancur DE, Kakiuchi M, Gupta S, et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 2020;581:100–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes & development 2013;27:1447–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenfeldt MT, O'Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature 2013;504:296–300 [DOI] [PubMed] [Google Scholar]
- 38.Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer discovery 2013;3:1272–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie X, Koh JY, Price S, White E, Mehnert JM. Atg7 Overcomes Senescence and Promotes Growth of BrafV600E-Driven Melanoma. Cancer discovery 2015;5:410–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santanam U, Banach-Petrosky W, Abate-Shen C, Shen MM, White E, DiPaola RS. Atg7 cooperates with Pten loss to drive prostate cancer tumor growth. Genes & development 2016;30:399–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes & development 2011;25:1510–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang A, Herter-Sprie G, Zhang H, Lin EY, Biancur D, Wang X, et al. Autophagy Sustains Pancreatic Cancer Growth through Both Cell-Autonomous and Nonautonomous Mechanisms. Cancer discovery 2018;8:276–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poillet-Perez L, Xie X, Zhan L, Yang Y, Sharp DW, Hu ZS, et al. Autophagy maintains tumour growth through circulating arginine. Nature 2018;563:569–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 2016;536:479–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poillet-Perez L, Sharp DW, Yang Y, Laddha SV, Ibrahim M, Bommareddy PK, et al. Autophagy promotes growth of tumors with high mutational burden by inhibiting a T-cell immune response. Nat Cancer 2020;1:923–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White E, Lattime EC, Guo JY. Autophagy Regulates Stress Responses, Metabolism, and Anticancer Immunity. Trends Cancer 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bryant KL, Stalnecker CA, Zeitouni D, Klomp JE, Peng S, Tikunov AP, et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng J, Thennavan A, Dolgalev I, Chen T, Li J, Marzio A, et al. ULK1 inhibition overcomes compromised antigen presentation and restores antitumor immunity in LKB1 mutant lung cancer. Nat Cancer 2021;2:503–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer discovery 2014;4:914–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cassidy LD, Young ARJ, Young CNJ, Soilleux EJ, Fielder E, Weigand BM, et al. Temporal inhibition of autophagy reveals segmental reversal of ageing with increased cancer risk. Nat Commun 2020;11:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cassidy LD, Young AR, Perez-Mancera PA, Nimmervoll B, Jaulim A, Chen HC, et al. A novel Atg5-shRNA mouse model enables temporal control of Autophagy in vivo. Autophagy 2018;14:1256–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Gokhale S, Jung J, Zhu S, Luo C, Saha D, et al. Mitochondrial Fission Factor Is a Novel Interacting Protein of the Critical B Cell Survival Regulator TRAF3 in B Lymphocytes. Front Immunol 2021;12:670338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moore CR, Liu Y, Shao C, Covey LR, Morse HC 3rd, Xie P. Specific deletion of TRAF3 in B lymphocytes leads to B-lymphoma development in mice. Leukemia 2012;26:1122–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gokhale S, Lu W, Zhu S, Liu Y, Hart RP, Rabinowitz JD, et al. Elevated Choline Kinase alpha-Mediated Choline Metabolism Supports the Prolonged Survival of TRAF3-Deficient B Lymphocytes. J Immunol 2020;204:459–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lalani AI, Moore CR, Luo C, Kreider BZ, Liu Y, Morse HC 3rd, et al. Myeloid cell TRAF3 regulates immune responses and inhibits inflammation and tumor development in mice. J Immunol 2015;194:334–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, et al. Glucose feeds the TCA cycle via circulating lactate. Nature 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dow LE, Premsrirut PK, Zuber J, Fellmann C, McJunkin K, Miething C, et al. A pipeline for the generation of shRNA transgenic mice. Nature protocols 2012;7:374–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hensley CT, DeBerardinis RJ. In vivo analysis of lung cancer metabolism: nothing like the real thing. The Journal of clinical investigation 2015;125:495–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sellers K, Fox MP, Bousamra M 2nd, Slone SP, Higashi RM, Miller DM, et al. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. The Journal of clinical investigation 2015;125:687–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab 2016;23:517–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roy S, Leidal AM, Ye J, Ronen SM, Debnath J. Autophagy-Dependent Shuttling of TBC1D5 Controls Plasma Membrane Translocation of GLUT1 and Glucose Uptake. Molecular cell 2017;67:84–95 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karim S, Adams DH, Lalor PF. Hepatic expression and cellular distribution of the glucose transporter family. World J Gastroenterol 2012;18:6771–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.TeSlaa T, Bartman CR, Jankowski CSR, Zhang Z, Xu X, Xing X, et al. The Source of Glycolytic Intermediates in Mammalian Tissues. Cell Metab 2021;33:367–78 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Z, Dong C. Gluconeogenesis in Cancer: Function and Regulation of PEPCK, FBPase, and G6Pase. Trends Cancer 2019;5:30–45 [DOI] [PubMed] [Google Scholar]
- 65.Icard P, Wu Z, Alifano M, Fournel L. Gluconeogenesis of Cancer Cells Is Disrupted by Citrate. Trends Cancer 2019;5:265–6 [DOI] [PubMed] [Google Scholar]
- 66.Grasmann G, Smolle E, Olschewski H, Leithner K. Gluconeogenesis in cancer cells - Repurposing of a starvation-induced metabolic pathway? Biochim Biophys Acta Rev Cancer 2019;1872:24–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Romero Vielva L Tumor lymphocytic infiltration in non-small cell lung cancer: the ultimate prognostic marker? Transl Lung Cancer Res 2016;5:370–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yan X, Jiao SC, Zhang GQ, Guan Y, Wang JL. Tumor-associated immune factors are associated with recurrence and metastasis in non-small cell lung cancer. Cancer Gene Ther 2017;24:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khayati K, Bhatt V, Hu ZS, Fahumy S, Luo X, Guo JY. Autophagy compensates for Lkb1 loss to maintain adult mice homeostasis and survival. Elife 2020;9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Faubert B, Li KY, Cai L, Hensley CT, Kim J, Zacharias LG, et al. Lactate Metabolism in Human Lung Tumors. Cell 2017;171:358–71 e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dufour JH, Dziejman M, Liu MT, Leung JH, Lane TE, Luster AD. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J Immunol 2002;168:3195–204 [DOI] [PubMed] [Google Scholar]
- 72.Sgadari C, Angiolillo AL, Cherney BW, Pike SE, Farber JM, Koniaris LG, et al. Interferon-inducible protein-10 identified as a mediator of tumor necrosis in vivo. Proceedings of the National Academy of Sciences of the United States of America 1996;93:13791–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arenberg DA, Kunkel SL, Polverini PJ, Morris SB, Burdick MD, Glass MC, et al. Interferon-gamma-inducible protein 10 (IP-10) is an angiostatic factor that inhibits human non-small cell lung cancer (NSCLC) tumorigenesis and spontaneous metastases. The Journal of experimental medicine 1996;184:981–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fujimoto Y, Inoue N, Morimoto K, Watanabe T, Hirota S, Imamura M, et al. Significant association between high serum CCL5 levels and better disease-free survival of patients with early breast cancer. Cancer Sci 2020;111:209–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoshimura T. The chemokine MCP-1 (CCL2) in the host interaction with cancer: a foe or ally? Cell Mol Immunol 2018;15:335–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data for this study were generated at Immune monitoring core facility and Metabolic core facilty at Rutgers Cancer Institute of New Jersey. Derived data supporting the findings of this study are available from the corresponding author upon request.