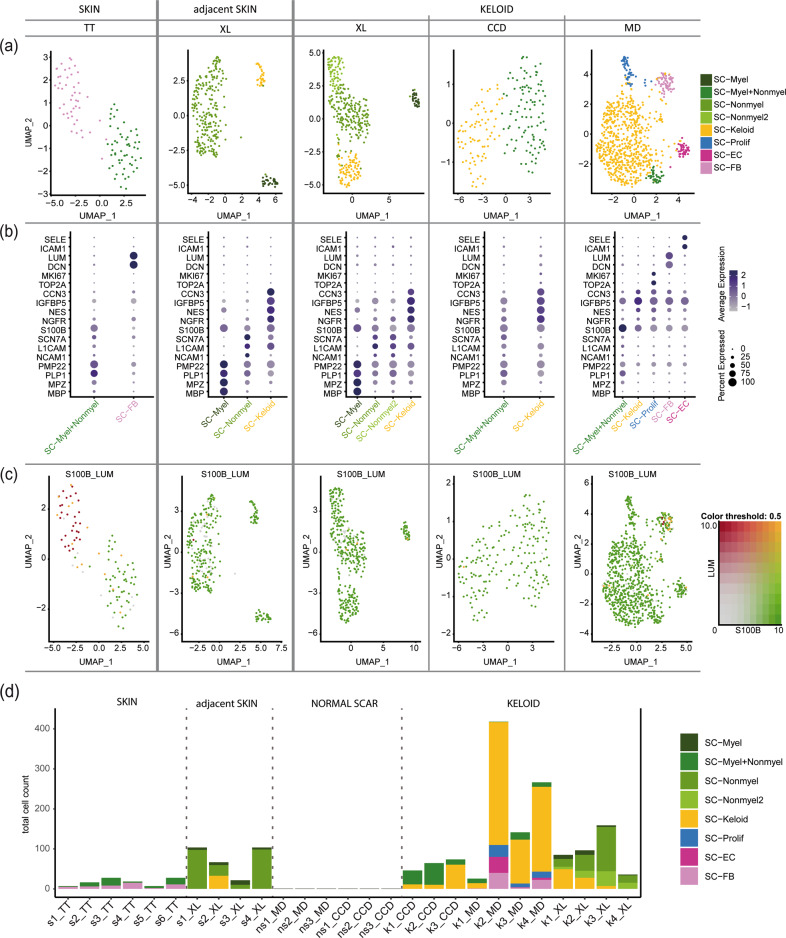
Fig. 4. Tissue- and source-specific dataset evaluation confirms keloidal profibrotic Schwann cells.
a UMAP plots depict subsets of identified Schwann cell clusters. Data sources: Tracy Tabib et al., 2018 (TT)18; Xuanyu Liu et al., 2021 (XL)15, Cheng-Cheng Deng et al., 2021 (CCD)16, Martin Direder et al., 2021 (MD)17; b Dot plots of well-known marker genes to characterize Schwann cell subtypes. MBP, MPZ, PLP1, and PMP22 for myelinating Schwann cells (SC-Myel); NCAM1, L1CAM, and SCN7A for nonmyelinating Schwann cells (SC-Nonmyel, SC-Nonmyel2); S100B and NGFR as general Schwann cell markers; NES, IGFBP5, CCN3 for keloidal Schwann cells (SC-Keloid); TOP2A and MKI67 for proliferating Schwann cells (SC-Prolif), DCN and LUM for cells expressing Schwann cell and fibroblast-specific genes (SC-FB), ICAM1 and SELE for cells expressing Schwann cell and endothelial cell-specific genes (SC-EC); mixed myelinating and nonmyelinating Schwann cell cluster (SC-Myel+Nonmyel); Color codes indicate average gene expression levels; Dot sizes visualize relative amounts of positive cells. c Feature blends reveal cells positive for S100B (green) and LUM (red) and double-positive cells (yellow). d Bar plots show the absolute cell count of distinct Schwann cell clusters in each included dataset. skin (s), normal scar (ns), keloid (k); data source: Tracy Tabib et al., 2018 (TT)18; Xuanyu Liu et al., 2021 (XL)15, Cheng-Cheng Deng et al., 2021 (CCD)16, Martin Direder et al., 2021 (MD)17.