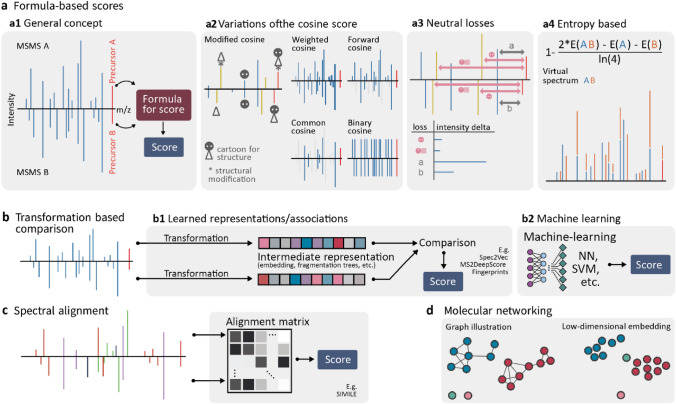
Fig. 1.
Overview of different spectral comparison (a–c) and spectral organisation methods (d) for two MS/MS spectra A and B. a1) Using mass spectral binning (i.e., to account for small m/z value differences), mass fragmentation spectra are transformed into vectors that are subsequently compared using mathematical formulas. a2) Modifications of the binning schema can account for other differences than m/z values (e.g., account for neutral losses, use only fragments present in both spectra, etc.). a3) Besides the actual mass fragment signals, neutral losses within or between spectra alone can serve as input for the spectral comparisons. a4) The Entropy score is a recently developed and high-performing metric for spectral comparisons. b1) Spectral comparison can be based on automatically computer-learned representations (i.e., alternatives to fragment spectral binning). b2) Comparison of MS/MS spectra can be achieved automatically with machine/deep learning methods and thus correlate better with structural similarity (NN: Neural Networks, SVM: Support Vector Machines). c Fragment spectra can be “aligned” similar to sequence alignment, which will report sub-spectra with overlapping fragments (i.e., certain structure parts of the two molecules, SIMILE: Significant Interrelation of MS/MS Ions via Laplacian Embedding). d Many MS/MS spectra can be organised into groups (molecular networking or mass spectral networking) or embedded in a lower subspace (a proxy for structural similarity)