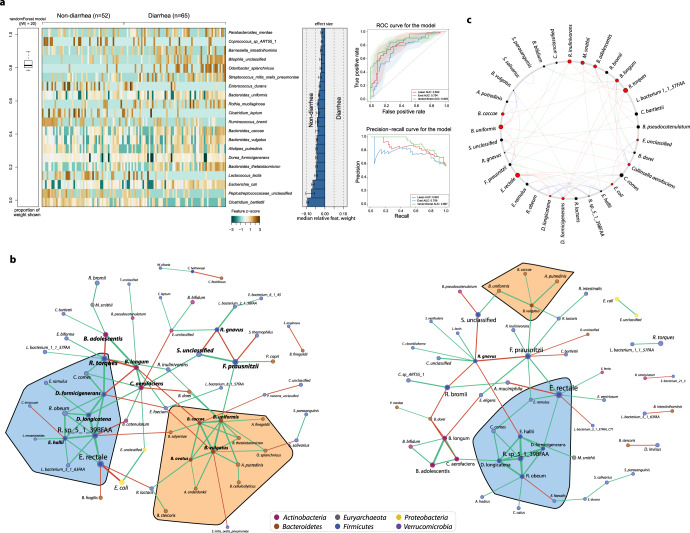
Fig. 3. Development of machine learning classifier that uses species abundances to distinguish between diarrheal and non-diarrheal sample types.
a The heatmap showing the relative abundances of top 20 discriminatory taxa between diarrheal and non-diarrheal PreTD types. Barplot depicts the effect size of the discriminatory taxa obtained from Random forest model. Cross-validation accuracy depicted as AUC-ROC and Precision-recall curve for the models built with three independent methods: ElasticNet (blue), Lasso (red) and Random Forest (green). b Co-occurrence network of diarrheal (left) and non-diarrheal (right) samples. The sizes of the nodes are proportional to the mean relative abundances of the species. The edges between two nodes represent significant correlations (green: positive; red: negative) between the two species. The orange highlighted region shows Bacteroidetes that are enriched in diarrhea (left) compared to non-diarrhea (right) network. The blue-highlighted region shows a conserved relationship among Firmicutes in both networks. c Circos plot depicting the common sub-network species between diarrheal and healthy sample networks. Node sizes are proportional to their scaled NESH score obtained from Netshift45, and species that gained connectivity in the diarrhea network compared to the healthy network are colored red. Connectivity changes between the common subnetworks are represented by connections between these species (red: connections that are unique to diarrhea samples network; blue: connections that are shared in both networks; green: connections that are unique to non-diarrheal samples network). Underlying data are provided in the Source Data file.