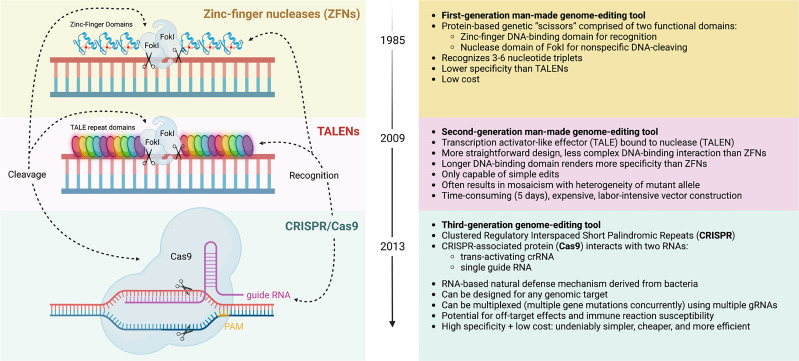
Figure 2.
Evolution of gene editing toolkits. Zinc-finger nucleases (ZFNs) are a first-generation man-made gene-editing tool that consists of two domains: 1) a DNA-cleaving domain comprised of the non-specific nuclease domain of FokI to introduce DNA double-stranded breaks, and 2) two DNA-binding domain chains, called “finger” modules, each recognizing a unique hexamer (6 bp) sequence of DNA. The fusion of DNA-binding and DNA-cleaving domains forms a pair of “genomic scissors”, with high specificity but lower than that of Transcription activator-like effector nucleases (TALENs) or Clustered Regulatory Interspaced Short Palindromic Repeats (CRISPR) gene editing systems. TALENs are a second-generation gene-editing tool also based on chimeric nucleases like ZFN with the same non-specific DNA cleavage domain (FokI), but it comprises of longer sequence-specific DNA-binding TALE modules, each of which contacts a single DNA base pair. By fusing with nuclease (TALE-Nuclease or TALEN), this tool can be used to edit genes, but only one at a time, unlike CRISPR. The longer programmable DNA-binding domain comprises of a series of 33-35 amino acid repeat domains, which allows for improved specificity than ZFNs, but increases cost and is marked by a low-efficiency process of vector construction. TALEN editing may induce mosaicism, in which mutations are only present in some transfected cells. The advent of CRISPR/Cas9 gene editing more recently has allowed for the creation of a tool with low cost, high specificity, high efficiency, and much simpler construction. This RNA-based tool is derived from a natural bacterial defense mechanism. The nuclease consists of the CRISPR-associated protein (Cas9) protein classically, though other Cas proteins have been used for cleaving. The complex initially binds to a short sequence known as the protospacer adjacent motif (PAM). This nuclease interacts with two RNAs for recognition and specificity, a trans-activating crRNA and single guide RNA, sometimes combined for simplicity. Notably, CRISPR allows for multiplexed gene editing, where multiple gene mutations can be issued concurrently. Complications include the potential for off-target effects and potential immune reaction susceptibility. As indicated, all systems employ a recognition module and cleavage domain that can be manipulated independently. Created with BioRender.com.