Extended Data Fig. 6 |. Target analysis for miR-21, miR-34a, and miR-324.
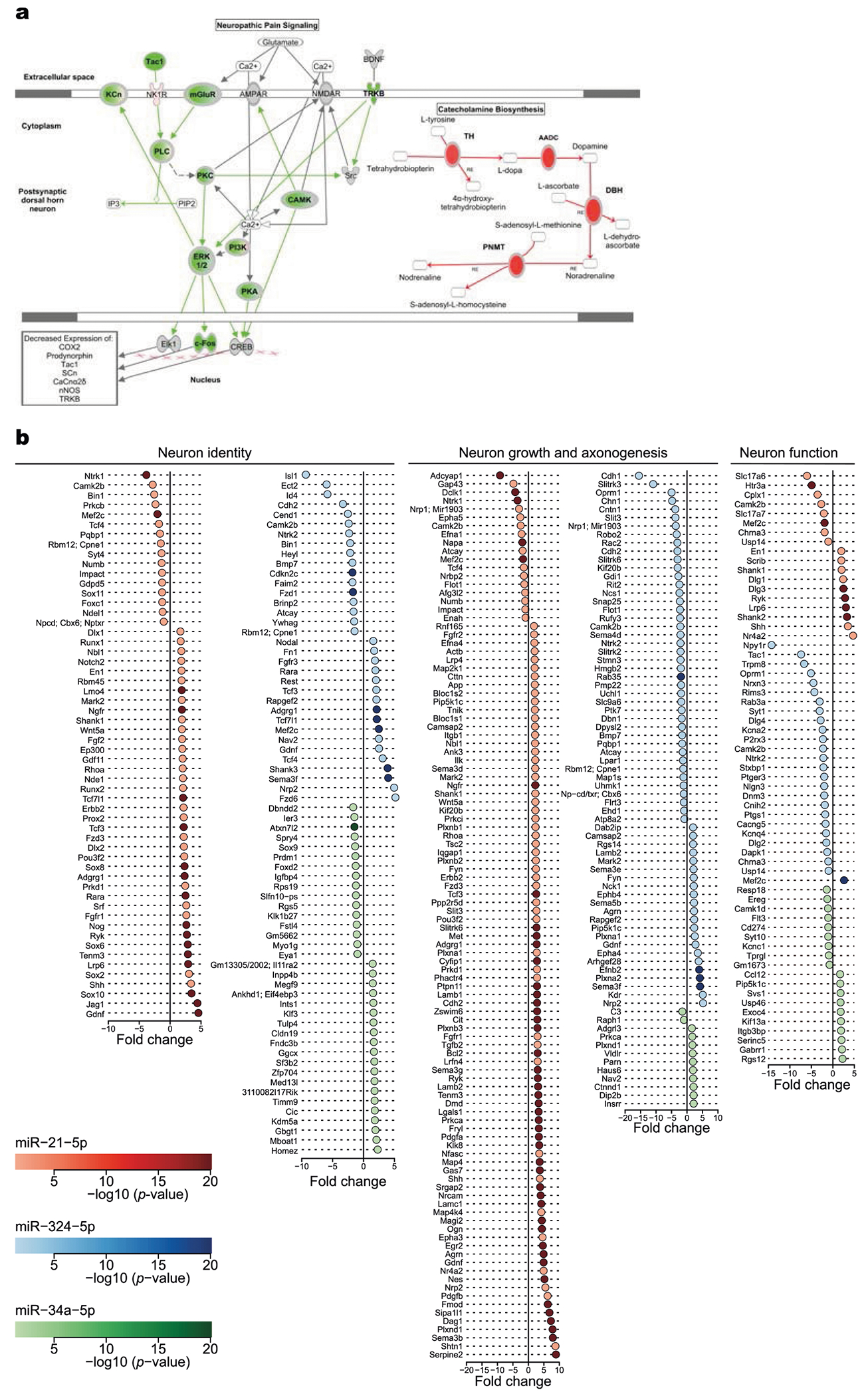
a, Schematic of ingenuity pathway analysis of pain signalling (silenced, green) and noradrenaline biosynthesis (activated, red). AADC, aromatic l-amino acid decarboxylase; DBH, dopamine β-hydroxylase; PNMT, phenylethanolamine N-methyltransferase. b, Total RNA was collected to assess global gene changes after TG neurons were transfected with miR-21 (red), miR-324 (blue), miR-34a (green), or scrambled miRNA. Data show fold change in expression of potential targets involved in neural identity determination (left), neural growth (middle), and neural function (right) between neurons transfected with different miRNAs (n = 3 biologically independent samples per condition). P values obtained from the moderated t-statistic for the presented genes were <0.05. Forest plots displaying fold change and P values for genes on different neuron pathways that are significantly differentiated between TG neurons transfected with miR-21, miR-34a or miR-324 and scramble miRNA. Linear models and empirical Bayes methods were used for obtaining the statistics and assessing differential gene expression between two conditions.
