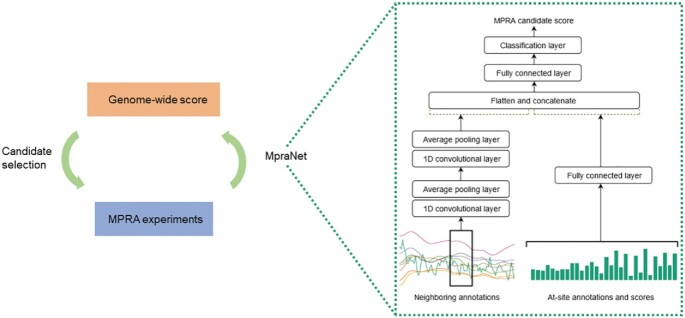
Figure 1.
MpraNet Architecture. The MpraNet model takes as input at-site functional annotations and scores as well as neighboring epigenetic annotations. The neighboring annotations are processed by 1D convolutional layers while at-site scores are processed by fully connected layers. The outputs from these networks are concatenated and passed into the final fully connected layers for classification. The model is run across the genome to produce genome-wide scores, which can be used to prioritize variants for future MPRA. Validated variants can be added to the MpraNet training set to further improve model performance.