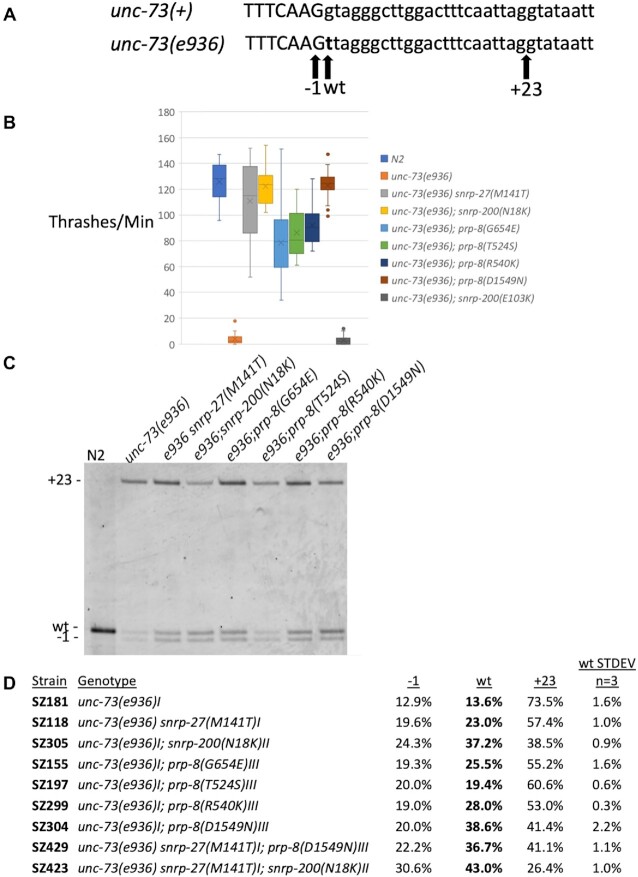
Figure 1.
New extragenic splicing suppressors of unc-73(e936). (A) Schematic of the unc-73 exon 16/intron 16 boundary. The top line shows the wildtype sequence and the bottom line shows the e936 G→T point mutation at the start of intron 16 in bold. Positions of the −1, wt and +23 cryptic 5′ splice site intron starts are indicated by arrows below. (B) A thrash test assay to demonstrate phenotypic suppression of unc-73(e936) uncoordination by the new suppressor allele mimics generated by CRISPR. The number of movements across the body axis per minute in liquid were measured for 20 different L4s for each indicated strain. (C) Reverse transcription and polymerase chain reaction (RT-PCR) analysis of unc-73(e936) splicing in the presence of different extragenic suppressors. Genotype of each strain is indicated above each lane (N2 is the wild isolate Bristol N2 strain), and the bands corresponding to the different cryptic splice sites are shown on the left. The reverse PCR primer was 5′ end labeled with Cy3, and the products separated on a 6% acrylamide denaturing gel and visualized using a Typhoon scanner. The three new suppressor alleles described in this manuscript are snrp-200(N18K), prp-8(R540K) and prp-8(D1549N). Previously identified class II suppressors snrp-27(M141T) (16), prp-8(G654E) and prp-8(T524S) (17) are shown here for comparison. (D) Quantitation of RT-PCR products for unc-73(e936) for the indicated strains. The mean value and standard deviation of three separate experiments are shown (see Supplementary Table S1 for experimental values and P-values relative to the unsuppressed strain). Note that in these class II suppressors, +23 splicing is decreased and −1 and wt site usage are both increased.