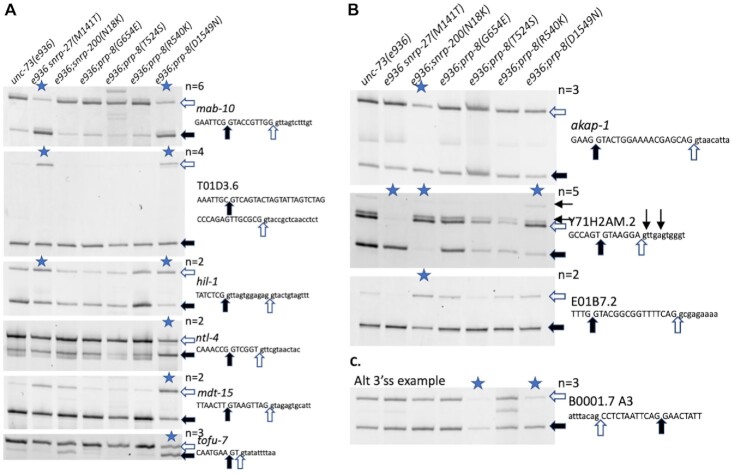
Figure 2.
Reverse transcription and polymerase chain reaction (RT-PCR) analysis of RNA from unc-73(e936) suppressor strains on selected alternatively spliced introns. Sequence of the 5′ss (A and B) or 3′ss (C) region of the alternative spliced gene is indicated at the right and filled and clear arrows indicate alternative splice sites and correspond to the bands indicated on the scanned gels. Samples with a mean ΔPSI >20% compared to the unc-73(e936) unsuppressed control strain are indicated with a blue star at the top of the lane; the number of independent trials used to calculate mean ΔPSI (n) is indicated. Quantitation data for these experiments is provided in Supplementary Table S1. (A) Sample alternative 5′ splicing events with changes in the prp-8(D1549N) mutant strain. Note in the first three examples that the snrp-27(M141T) mutant strain has similar effects on alternative splicing while in the next three examples the effect is specific to D1549N. (B) Sample alternative 5′ splicing events from the snrp-200(N18K) mutant strains. Note that for the Y71H2AM.2 intron shown, the prp-8(D1549N) mutation has a similar effect on alternative 5′ss choice, while the snrp-27(M141T) mutation has the dramatic opposite effect. Two minor confirmed alternative 5′ss for Y71H2AM.2 are indicated above the sequence with small arrows. (C) An example of an alternative 3′ splicing event uncovered in the prp-8(D1549N) RNASeq analysis. Note that the change in alternative 3′ss usage seen for this intron in the prp-8(T524S) suppressor strain was reported previously (17).