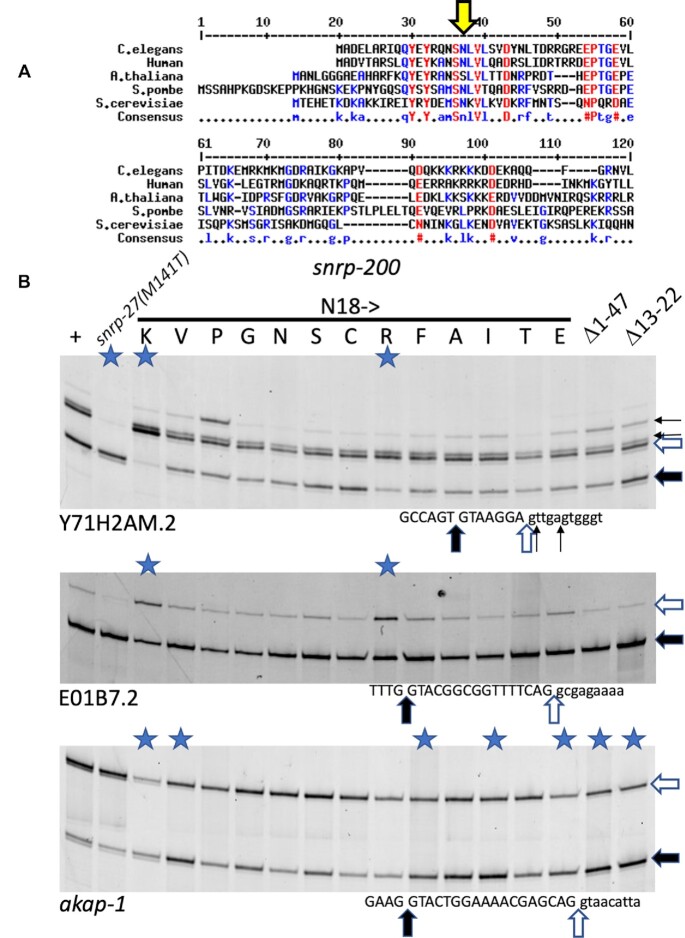
Figure 6.
Alternative 5′ splicing tests on a range of SNRP-200(N18) substitutions. (A) Alignment of the N-terminal region of SNRNP200/BRR2 from C. elegans, human, Arabidopsis thaliana, the fission yeast Schizosaccharomyces pombe and S. cerevisiae. The large yellow arrow indicates the N18 residue in C. elegans. (B) RT-PCR analysis of mutants on specific introns. The first lane on the left is from RNA from wild type strain N2. The second lane uses RNA derived from a strain with a mutation in snrp-27(M141T). The next 13 lanes use RNA derived from strains generated by CRISPR mutagenesis at the N18 position; the amino acid substitutions at position N18 are indicated. Note that we recovered an N18N substitution by CRISPR, and that is a useful control in the seventh lane. The last two lanes use RNA derived from strains with N-terminal deletions of SNRP-200, amino acids 1–47 or amino acids 13–22 respectively. The 5′ splice site regions of the three introns, from Y71H2AM.2, E01B7.2 and akap-1 are indicated below each gel image. Lanes in which the products show ΔPSI > 0.2 compared to wild type in two biological replicates are indicated with a blue star (see Supplementary Table S1 for quantitation).