Figure 7.
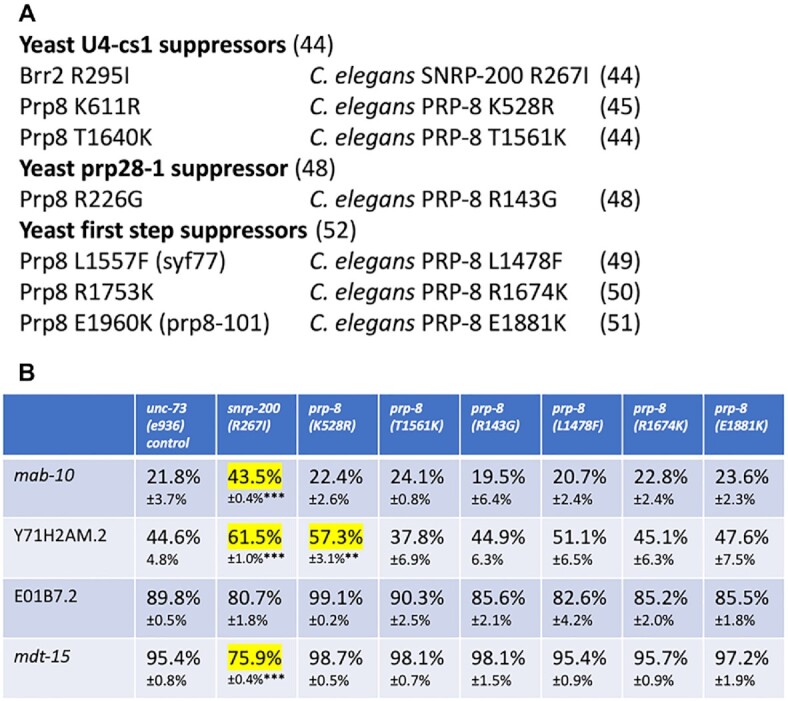
Generation and testing of yeast splicing suppressors of known classes in C. elegans. (A) The three different classes of yeast suppressor tested, the location of the seven individual alleles tested (with their location in S. cerevisiae BRR2 and PRP8 on the left and the homologous position of the suppressor in C. elegans on the right. Corresponding citations for the individual alleles and classes are indicated in parentheses. (B) Quantitation of alternative splicing of several introns regulated by class II suppressors in C. elegans strains bearing mimics of the yeast suppressors compared to an unsuppressed control in the left column. Percent usage of the upstream 5′ss and standard deviation (n = 3) are indicated. Events with >10% changes in upstream 5′ss usage relative to the control strain are highlighted in yellow. For the ones highlighted we determined the P-value for the change relative to the control. ** indicates P-value from student's t-test <0.01 and *** indicates P-value <0.001.
