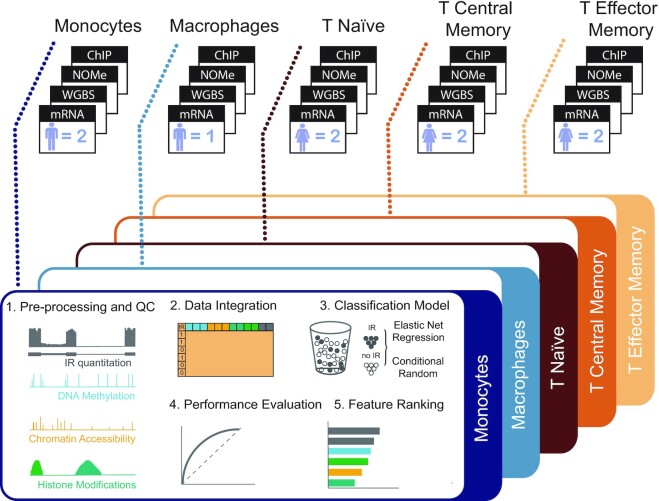
Figure 1.
Experimental design and computational workflow to determine regulators of IR. Raw high-throughput data were processed for each biological replicate and amalgamated by cell type from the indicated number of samples (n). The output was used for feature extraction: IR events were treated as a binary outcome and we trained an elastic net regression model and a conditional random forest model with a total of 48 sequence-based and epigenetic features. Using feature ranking, we identified the factors that were most strongly associated with IR outcomes and compared the performances of both modelling strategies. These steps were repeated for each cell type.