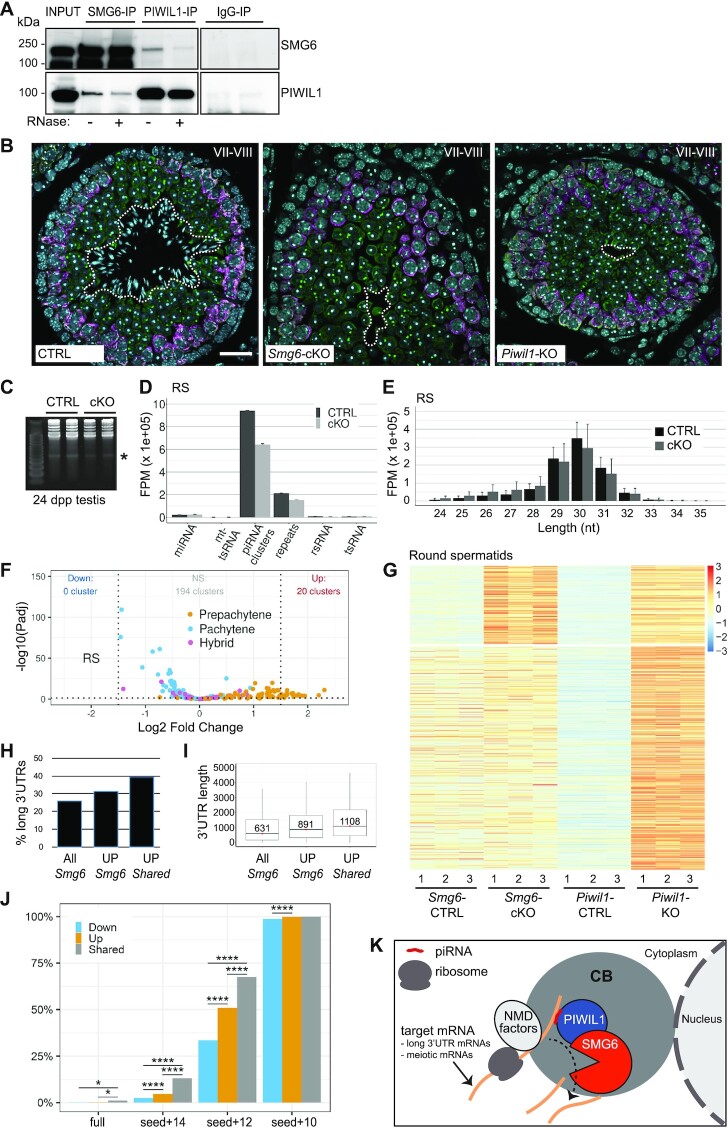
Figure 7.
Interplay of SMG6 with the piRNA pathway. (A) Co-IP of SMG6 and PIWIL1 from mouse testis with or without RNase treatment (+/−), followed by western blotting with antibodies against SMG6 and PIWIL1. Rabbit IgG was used as a negative control for IP. (B) IF of PFA-fixed paraffin embedded testis sections from control (CTRL), Smg6-cKO and Piwil1-KO mice. Antibodies against DDX4 (green) and PIWIL2 (magenta) were used to visualize the layers of PIWIL2-positive pachytene spermatocytes and DDX4-positive CB-containing round spermatids at stage VII–VIII. DAPI stains the nuclei. Dashed white line indicates the border between the layers of round spermatids and elongating spermatids. Scale bar: 25 μm. (C) Visualization of piRNAs in 24 dpp Smg6-cKO testes. Total RNA from CTRL and Smg6-cKO testes was run into a denaturating 15% polyacrylamide gel and stained with SYBRCold. piRNA band (around 30 nucleotides) is indicated. (D) Distribution of small-RNA-seq reads in genomic regions corresponding to miRNAs, mitochondrial tsRNAs (mt-tsRNA), piRNA clusters, repeats, ribosomal RNAs (rsRNA), and tsRNAs in control and Smg6-cKO round spermatids. Error bars represent mean ± SD of three biological replicates. (E) Size distribution of piRNA-mapping reads in round spermatids. Raw counts were normalized in FPM. Error bars represent mean ± SD of three biological replicates. FPM values for (D) and (E) were calculated using a robust median ratio method using DESeq2 v1.30.0. (F) Differential expression of piRNA clusters (small-RNA-seq) in round spermatids. piRNA clusters are divided into pre-pachytene (84 clusters), hybrid (30 clusters), and pachytene (100 clusters) according to their abundance in juvenile testes from 10.5 to 17.5 dp (20). (G) Heatmap showing all genes upregulated in Piwil1-KO round spermatids clustered according to their expression pattern in Smg6-cKO germ cells. Piwil1-KO transcriptome data was downloaded from NCBI GEO database (GSE42004). (H) The proportion (%) of long 3′UTR-containing transcripts (3′UTR > 1500 nt) among all significant transcripts (All Smg6, 25.9% of 22302 transcripts; Padj ≤ 0.05), transcripts upregulated in Smg6-cKO round spermatids (UP Smg6, 31.4% of 5771 transcripts; Padj ≤ 0.05, log2FC ≥ 1), and transcripts upregulated in both Smg6-cKO and Piwil1-KO round spermatids (UP Shared, 39.3% of 244 transcripts). (I) Box plots show the median values of 3′UTR length with the first and third quartiles for the same transcripts, whiskers show the range within 1.5 IQR (interquartile range). Outliers were removed for better visualization. (J) Percentage of genes with piRNA targeting sequenced among genes that are downregulated (Down) or upregulated (Up) in Smg6-cKO round spermatids, or upregulated in both Smg6-cKO and Piwil1-KO round spermatids (Shared). Pairing rules in target recognition: full = piRNA fully complementary to target mRNA, seed + 14, 12 or 10 = piRNA seed region fully complementary to target RNA plus additional 14, 12 or 10 nucleotide matches after the seed region. **P < 0.01; ****P < 0.0001, Fisher's exact test. (K) Model. NMD-recognized SMG6 target mRNAs are transported to the CB. piRNA-bound PIWIL1 participates in the recognition of SMG6-dependent NMD targets. See also Supplementary Figures S6 and S7, and Supplementary Tables S5 and S6.