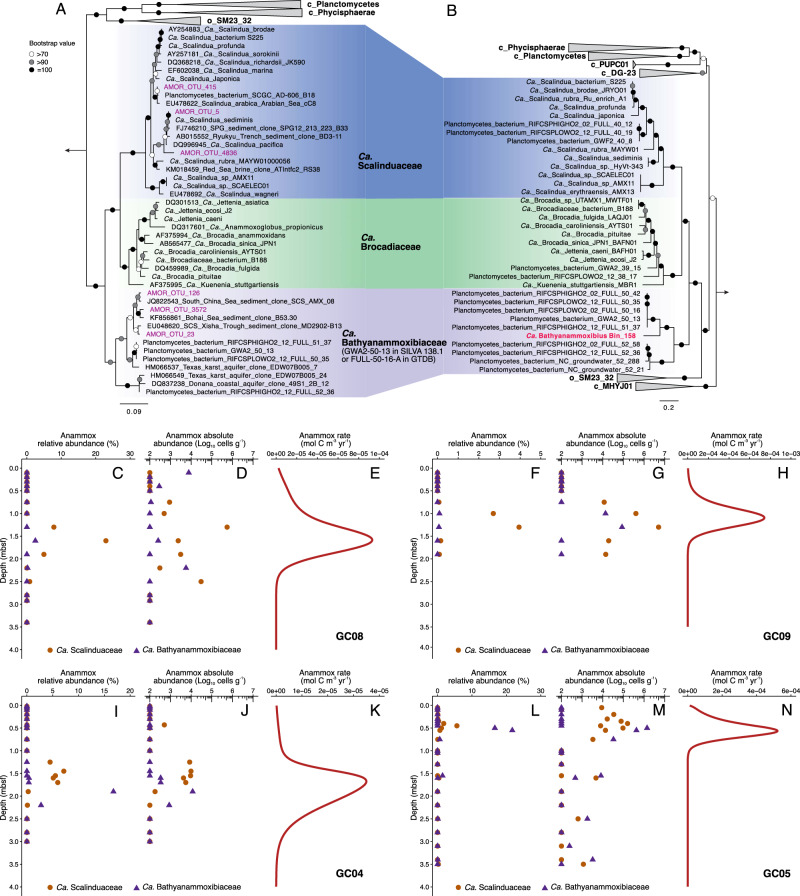
Fig. 1. Phylogeny and distribution of anammox bacteria in four AMOR sediment cores.
(A, N) Maximum-likelihood phylogenetic trees of bacteria in the order of Brocadiales based on the 16S rRNA gene (A) and 120 concatenated bacterial single-copy genes (B). The trees were inferred using IQ-TREE with GTR + F + R5 and LG + F + R7 as the best-fit evolutional model, and 1,000 times of ultrafast bootstrap iteration were applied to assess the robustness of both trees. The metagenome-assembled genome (MAG) recovered from the AMOR sediment is highlighted in red. The nomenclature used here following GTDB (https://gtdb.ecogenomic.org), while the new anammox family Ca. Bathyanammoxoceae (f__2-02-FULL-50-136 in GTDB 06-RS202 release) is named in this study. Bootstrap values of >70 are shown with symbols listed in the legend. The scale bars show estimated sequence substitutions per residue. C–N Vertical distribution of anammox bacteria and the predicted anammox reaction rates in AMOR sediment cores. The relative abundance of the two anammox families (B, E, H, K), Ca. Scalinduaceae and Ca. Bathyanammoxoceae, were determined by the sum of the OTUs assigned to these groups (showed in A), based on the 16S rRNA gene amplicon sequencing data reported in ref. [20]). The anammox reaction rates (D, G, J, M) were estimated by the reaction-transport model by ref. [20]. Panels (C–N) are modified from Fig. 2 of Ref. [20], an open access article distributed under Creative Commons Attribution License 4.0 (CC BY).