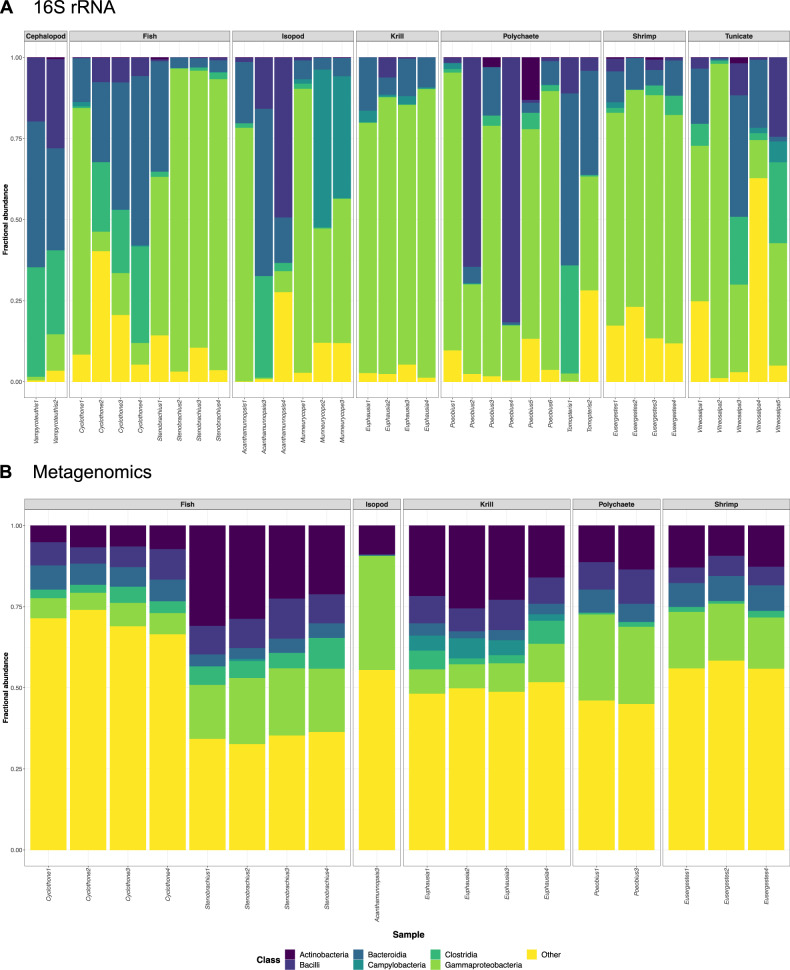
Fig. 1. Taxonomic composition of mesopelagic host-associated microbiomes.
Fractional abundances of microbial classes in the microbiomes of mesopelagic animals based on A 16S rRNA amplicon data and B metagenomic data. Gammaproteobacteria, Bacteroidia, Bacilli and Clostridia were the most prevalent bacterial classes across microbiomes based on 16S rRNA gene profiling (see also Table 2). These classes were also recovered through shotgun metagenomic sequencing, although with lower abundance. By contrast, a markedly higher proportion of Actinobacteria was revealed in each sample through this method (paired t-test: t = –8.2128, p < 2.548e–07).