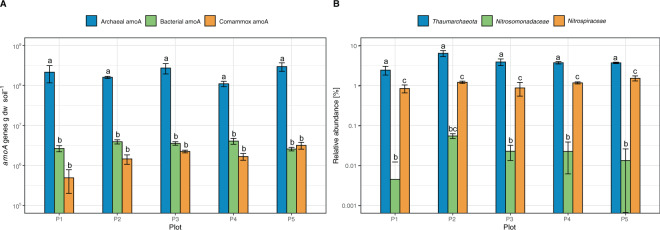
Fig. 3. Abundance of archaeal, canonical bacterial, comammox, and canonical nitrite-oxidzing bacteria in EAA soils based on amoA genes and relative 16S rRNA gene frequencies.
A Abundance of archaeal, canonical bacterial and comammox amoA genes in EAA soil plots 1 through 5 determined by qPCR. Error bars represent standard deviations of three sub-plots with three replicates each (n = 9). B Relative abundance of Thaumarchaeota, Nitrosomonadaceae, and Nitrospiraceae clades based on relative frequency of 16S rRNA genes in amplicon sequence datasets. Error bars represent standard deviations between three sub-plots (n = 3). Different letters indicates significant difference within plot. Archaeal amoA gene copy numbers were significant higher than bacterial and comammox amoA, differences between bacterial and comammox amoA were not statistically significant (ANOVA with Tukey HSD test at α = 0.05 level). Thaumarchaeota were significantly more frequent than Nitrosomonadaceae and Nitrospiraceae in all plots, Nitrospiraceae were significantly more frequent than Nitrosomonadacea, except in plot 2 (ANOVA with Tukey HSD test at α = 0.05 level).