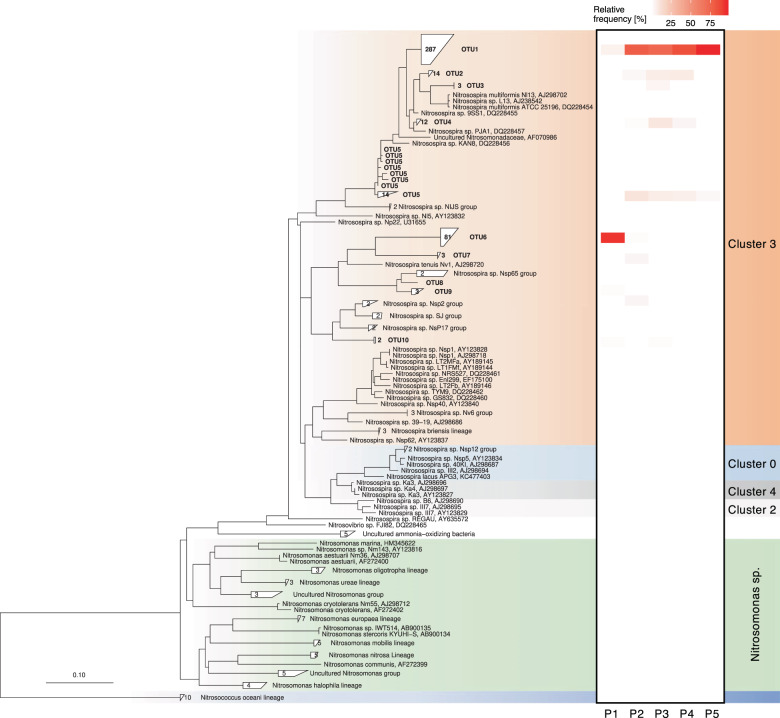
Fig. 5. Neighbor-joining phylogenetic tree of canonical bacterial amoA gene sequences retrieved from EAA plots 1 through 5 and heat map of relative abundance within each plot (in %).
Tree was calculated based on 453 aligned positions using neighbor-joining algorithm with Jukes–Cantor correction. Sequences with more than 97% sequence similarity on nucleotide level were grouped into single OTUs. The total number of sequences within a given OTU, if more than one, is indicated in each tree leaf. Labels on the right refer to Nitrosospira sp. sequence clusters 0–4.