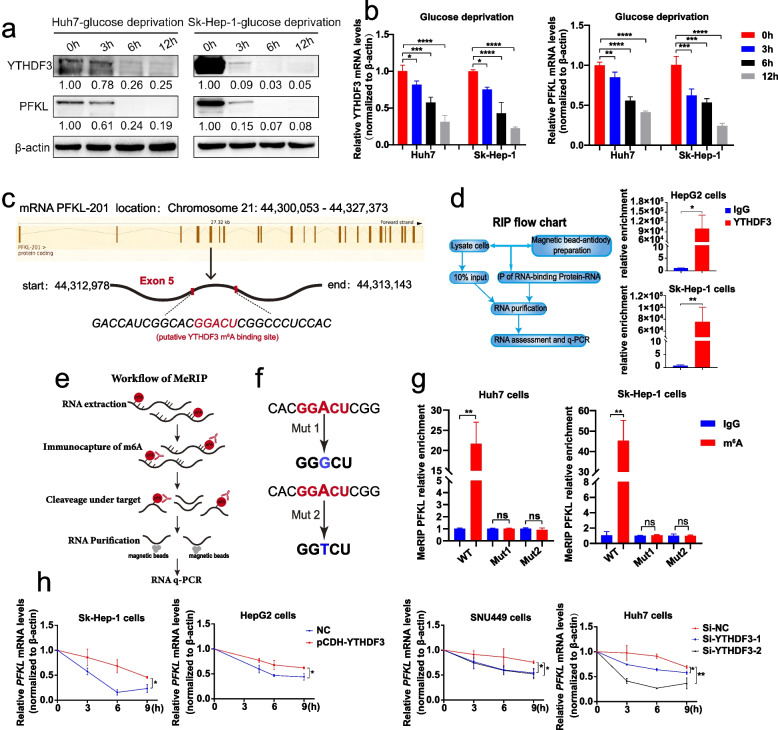
Fig. 7.
YTHDF3 suppressed the degradation of PFKL mRNA via m6A modification. a-b The relative mRNA and protein expression of YTHDF3 and PFKL under glucose deprivation condition for 0 h, 3 h, 6 h and 12 h in Huh7 and Sk-Hep-1 cells. (We cultured Huh7 and Sk-Hep-1 cells with glucose-free RPMI1640 medium for 0 h, 3 h, 6 h and 12 h). c Putative YTHDF3 m6A modified site to PFKL mRNA transcript according to information of SRAMP website. d The RIP- qPCR assay flow chat (left) and the relative enrichment folds of PFKL mRNA correlated with YTHDF3 in Sk-Hep-1 and HepG2 YTHDF3-stably-overexpressed cell lines (right). e Workflow of MeRIP- qPCR assay. f Two mutant PFKL plasmids used for MeRIP assays. Mut1: adenosine bases (A) in predicted m6A binding sites were replaced by guanine base (G). Mut2: adenosine bases (A) in predicted m6A binding sites were replaced by thymine base (T). g The relative enrichment folds of m6A modified PFKL mRNA by YTHDF3 with wild-type PFKL plasmid and two mutant PFKL plasmids in Sk-Hep-1 and Huh7 cells. h. PFKL mRNA stability analysis in HCC cells with YTHDF3 transiently knockdown or overexpression in the presence of actinomycin D (5 μg/ml) for 0 h, 3 h, 6 h and 9 h