Figure EV2. CCNL1 degradation requires Cul1 and is blocked when substrate‐binding deficient FBXW7R465C is co‐expressed.
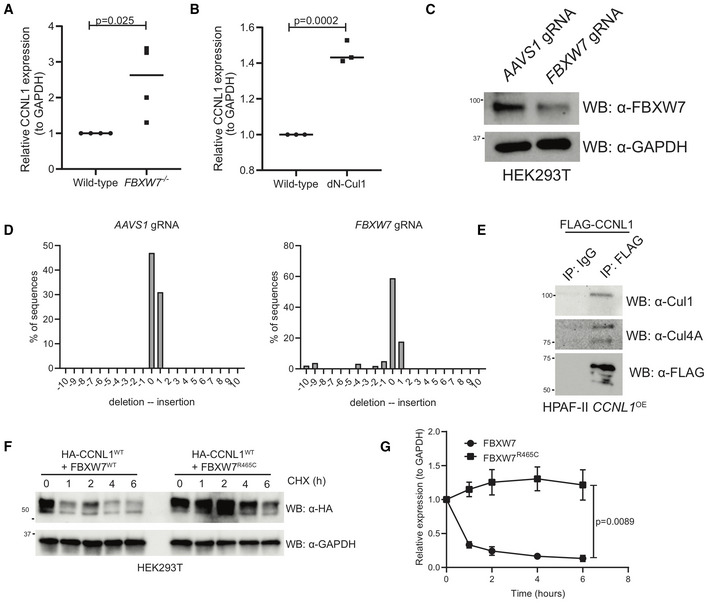
- Quantification of immunoblots in Fig 3A, mean ± SEM, students t‐test, n = 3 independent biological replicates.
- Quantification of immunoblots in Fig 3D, mean ± SEM, students t‐test, n = 3 independent biological replicates.
- Immunoblot analysis of HEK293T cells expressing gRNAs against AAVS1 or FBXW7.
- TIDE analysis of HEK293T cells expressing gRNAs against AAVS1 or FBXW7.
- Immunoblot of immunoprecipitation of FLAG‐CCNL1 overexpressed in HPAF‐II cells, detecting endogenous Cul1 and Cul4A. Representative image of three independent replicates.
- Immunoblot of lysates following cycloheximide treatment of HEK293T cells expressing HA‐CCNL1 and FLAG‐FBXW7 or FLAG‐FBXW7R465C. Representative blot of three independent replicates.
- Quantification of cycloheximide chase in (F), mean ± SEM of three independent replicates, t‐test at T6.
