Figure 1. Functional mapping of DDX60 antiviral domains and interrogation of anti‐HCV activity.
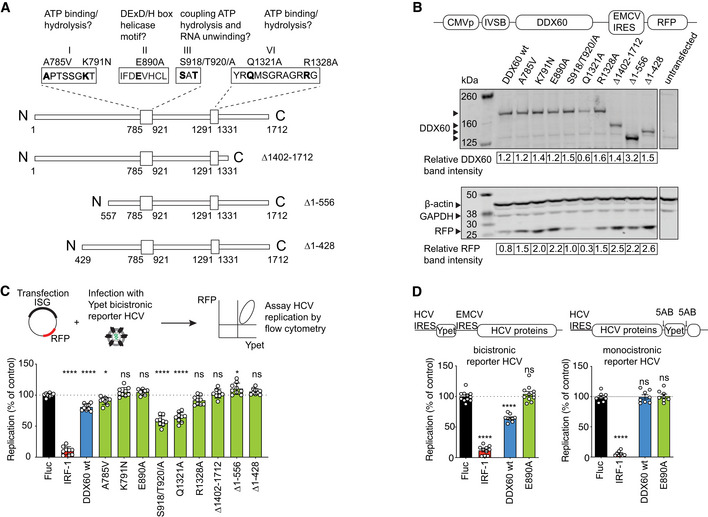
- Schematic of DDX60 protein with putative functional domains. Helicase ATP binding type I domain (amino acids 785–921) and C‐terminal helicase domain (amino acids 1,291–1,331) are shown as larger boxes in linear DDX60 schematic. Amino acids are numbered below. Putative functional motifs (I, II, III, and VI) and mutations made are annotated. The amino acids in bold as well as N‐ and C‐terminal regions were interrogated in antiviral assays.
- Assessment of exogenous DDX60 expression. HEK293T cells transfected with DDX60 wild‐type (wt), or DDX60 mutants and analyzed by Western blot for DDX60, ß‐actin and GAPDH (loading controls), and RFP (reporter). DDX60 and RFP quantification relative to GAPDH from one representative blot are shown below.
- HCV antiviral assays with DDX60 wt or mutant panel. Huh‐7 cells transfected with an RFP containing plasmid backbone encoding either Firefly luciferase (Fluc and negative control), IRF1 (positive antiviral control), DDX60 wt, or DDX60 mutants and infected with HCV‐Ypet, a bicistronic reporter HCV where Ypet reporter protein is driven by HCV IRES and HCV polyprotein consisting of C, E1, E2, p7, NS2, NS3, 4A, 4B, NS5A, and NS5B is driven by EMCV IRES.
- Effect of DDX60 on replication of bicistronic or monocistronic infectious reporter HCVs. Huh‐7 cells transfected as in (C) and infected with either bicistronic HCV‐Ypet (left) or monocistronic HCV J6/JFH‐5AB‐YPet. Ypet reporter in monocistronic HCV is placed in between NS5A and NS5B.
Data information: For (C) and (D), percent of Ypet+ cells in RFP+ cells is scaled to one replicate of Fluc control. Data shows mean ± SD for at least n = 3 biological replicates; ns —not significant, *P < 0.05, ****P < 0.0001, ns, nonsignificant using ANOVA and Dunnett's multiple comparison test against Fluc.
