Fig. 1. Hallucinating symmetric protein assemblies.
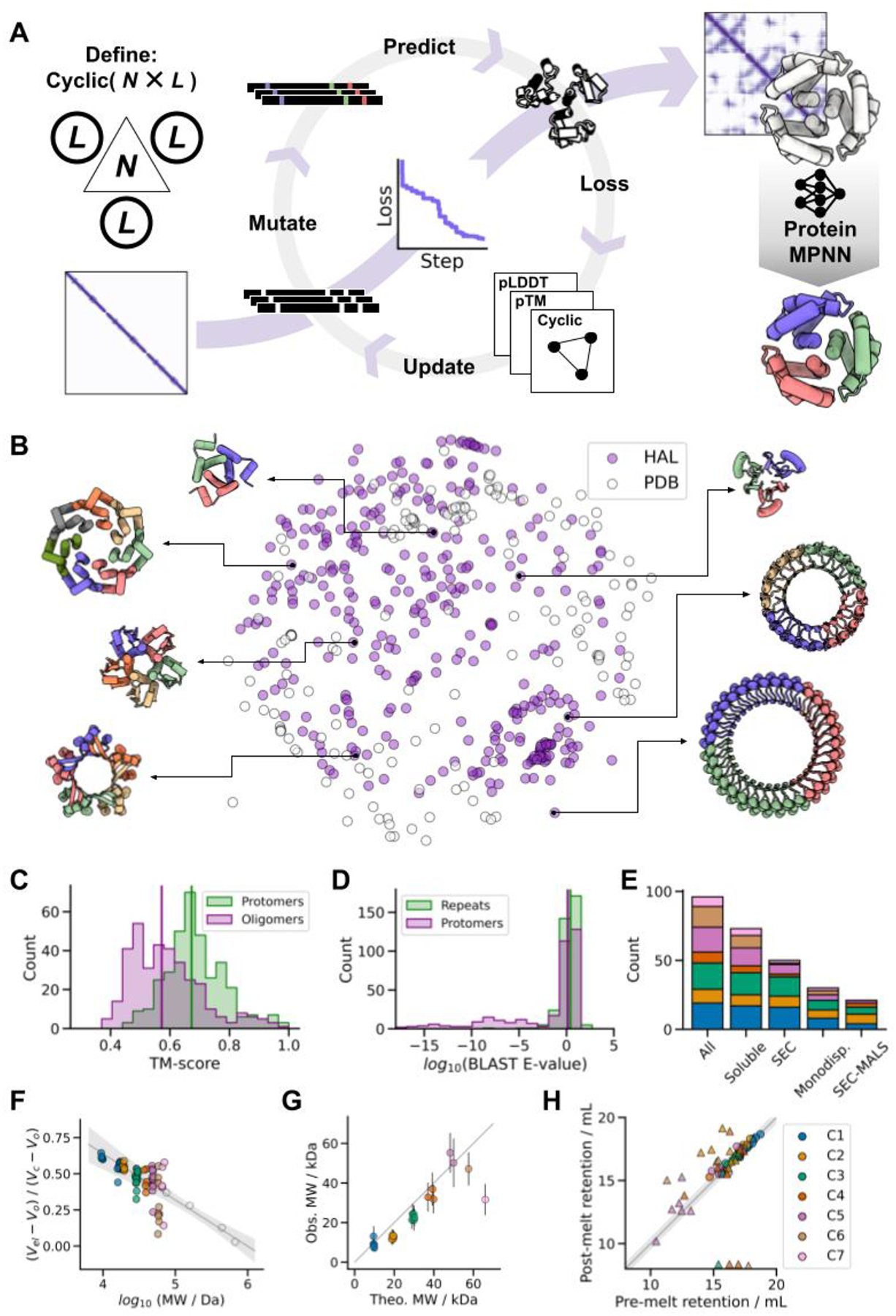
(A) Starting from choice of a cyclic symmetry and protein length, a random sequence is optimized by MCMC through the AF2 network until the resulting structure fits the design objective, followed by sequence re-design with ProteinMPNN. (B) The method generates structurally diverse outputs, quantified here by multi-dimensional scaling of protomer pairwise structural similarities between experimentally tested HALs (N = 351) and all de novo cyclic oligomers present in the PDB (N = 162). (C) Generated structures differ from those in the PDB. Median TM-scores to the closest match: 0.67 and 0.57 for the protomers and oligomers respectively (vertical lines). (D) Generated sequences are unrelated to naturally-occuring proteins. Median BLAST E-values from the closet hit in UniRef100: 2.6 and 1.3 for the repeat motifs and protomers respectively (vertical lines). (E) Success counts of ProteinMPNN-designed HALs at different levels of characterization. (F) Most soluble HALs have SEC retention volumes consistent with their oligomeric state. The gray line shows the fit to calibration standards (open circles), and the shaded area represents the 95% confidence interval of the calibration. (G) The observed molecular weights of HALs from SEC-MALS are close to those computed from the design models. (H) ProteinMPNN-designed HALs are thermostable. Pre-melting and post-melting retention volumes are closely correlated; circles represent designs that remained monodisperse, while triangles indicate polydispersity after heat-treatment. In plots E-H, the data is categorized by cyclic symmetry classes. The legend is shown in H.
