Figure 3 -. Integrative non-coding variant effect prediction.
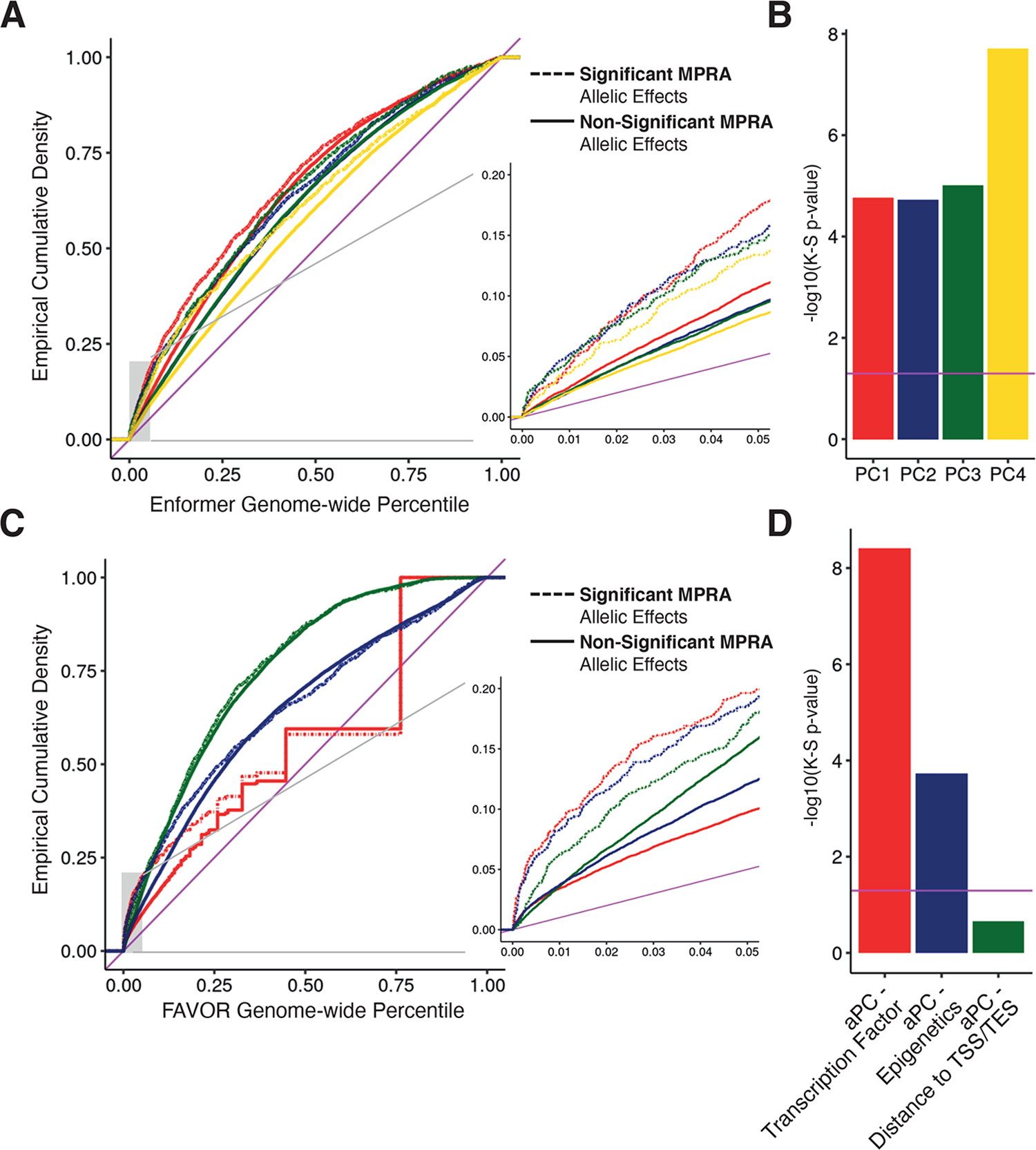
(A) Empirical cumulative probability distribution of the first through fourth principal component (PC) scores from Enformer for allelic MPRA hits and other tested variants significant and non-significant MPRA allelic hits; genome-wide percentiles computed across all common variants in 1000 Genomes Phase 3. Inset shows a blow up of lower genome-wide percentile curves (B) Significance of a Kolgomorov-Smirnov (K-S) test comparing the empirical distributions of Enformer scores for significant and non-significant allelic MPRA hits v; magenta horizontal line indicates significance by K-S test (p-value < 0.05). (C) Same as in (A) except showing annotation principle components from FAVOR; genome-wide percentiles computed across all variants in TOPMed Freeze5. (D) Same as in B, except testing FAVOR aPCs.
