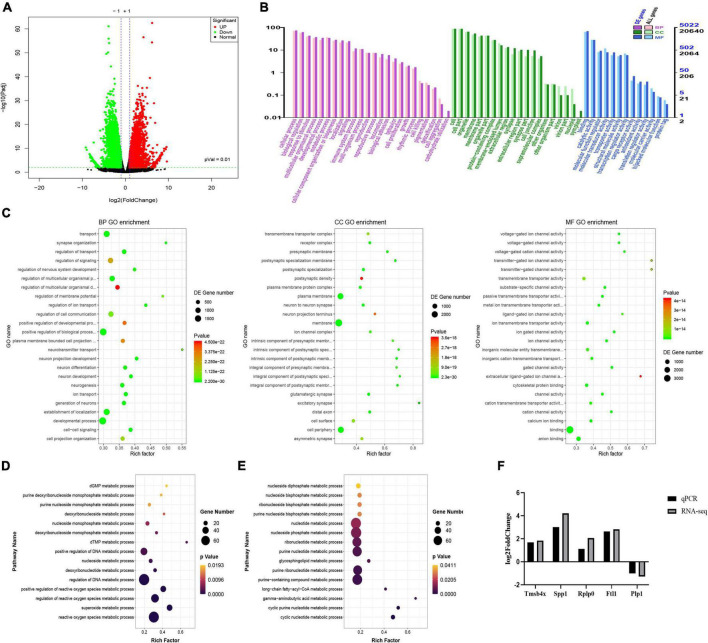
FIGURE 4.
Transcriptome changes during the acute-phase spinal cord injury. (A) Changes in genes between the sham and acute-phase spinal cord injury groups. Compared with the sham group, red indicates significant upregulation, green indicates significant downregulation, and black indicates no significant change in the acute-phase spinal cord injury, based on P < 0.01. (B) Gene ontology annotation classification statistics chart. The horizontal axis is the gene ontology classification, the vertical axis is the percentage of the number of genes on the left and the number of genes on the right. (C) The top 20 pathways enriched for differentially expressed genes in biological process, cellular component and molecular function are shown in order. (D) Metabolic processes involved in upregulated genes. The vertical axis is the name of the metabolic processes, and the horizontal axis is the number of genes annotated to that metabolic process and its number as a proportion of the total number of genes annotated on it. (E) Metabolic processes involved in downregulated genes. (F) Quantitative real-time polymerase chain reaction validation of transcriptome data. The result verified that the expression trends of the five genes selected randomly were consistent with the transcriptomic data, where Tmsb4x, Spp1, Rplp0, Ftl1 were upregulated and Plp1 was downregulated. GO, gene ontology; BP, biological process; CC, cellular component; MF, molecular function; QPCR, quantitative real-time polymerase chain reaction; Rplp0, ribosomal protein lateral stalk subunit P0; Tmsb4x, thymosin beta 4, X-linked; Ftl1, ferritin light chain 1; Spp1, secreted phosphoprotein 1; Plp1, proteolipid protein 1.