Figure 6. Nitrate reductase activity is required for efficient colonization in a mouse model of inflammation.
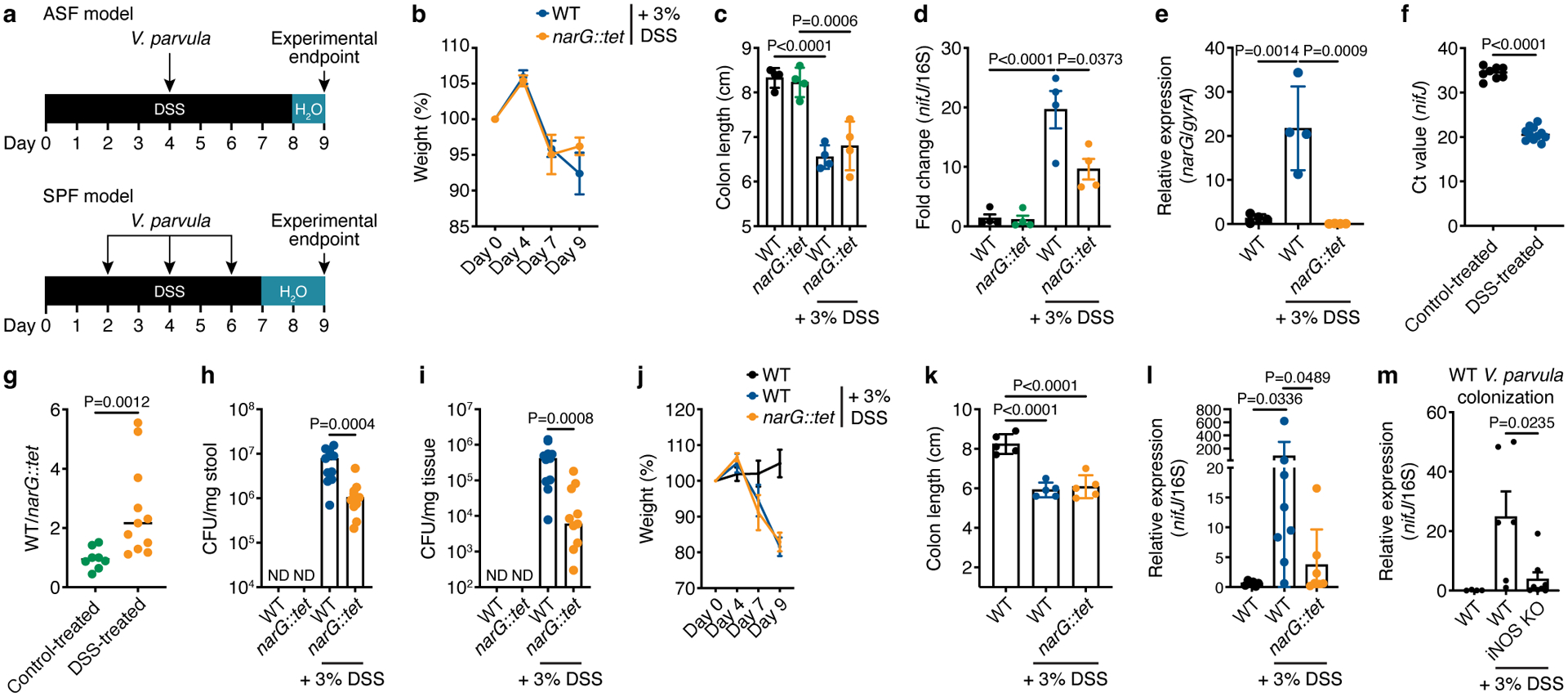
a) Experimental design of mouse models of colitis. Assessments of b) weight change over DSS treatment course and c) colon length at experimental endpoint (n=5). d) Relative abundance of Veillonella in stool of ASF mice, calculated using the ΔΔC(t) method comparing V. parvula nifJ and 16S rRNA genes (n=4). e) Relative expression of the narG gene in WT Veillonella colonizing stool of control- and DSS-treated ASF mice, with narG::tet-colonized mice serving as a negative control (n=4). gyrA was used as an internal control. f) Ct values of the Veillonella nifJ gene measured by qPCR in stool of WT-colonized ASF mice with and without DSS treatment (n=8–11). g) qPCR analysis of the relative colonization of WT and narG::tet strains in stool of ASF mice inoculated with a 1:1 mixture of each strain with and without DSS treatment (n=8–11). The y-axis represents the fold-change difference in abundance of each strain. Colony-forming units (CFUs) measured from h) stool and i) colonic tissue of ASF mice inoculated 1:1 with WT and narG::tet strains (n=8–11). ND, not detected. Weight change j) and colon length k) in SPF mice (n=5). l) Colonization of WT and narG::tet strains in stool of SPF mice measured by qPCR, comparing Ct values of nifJ and 16S rDNA (n=6–8). m) Colonization of WT Veillonella in stool of WT and iNOS KO DSS-treated SPF mice compared to control-treated WT mice measured by qPCR (n=4–8). Data represent two independent experiments and show mean (b-g, j-m) ± SEM (b-e, j-m) or median (h,i) values. Data was analyzed by one-way ANOVA and Tukey’s HSD test or Mann-Whitney U-test. NS, not significant. N numbers denote mice.
