FIG. 1.
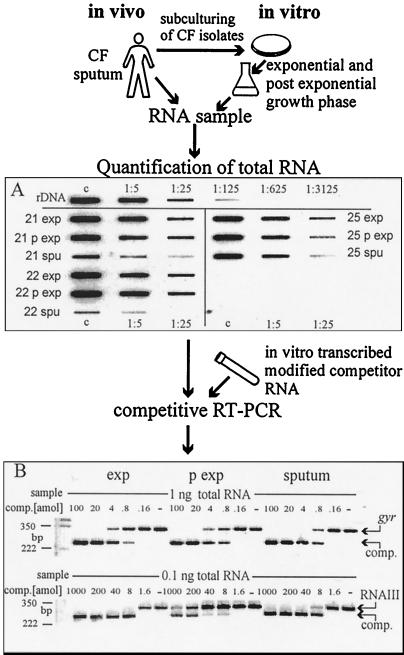
S. aureus cells were lysed nonenzymatically in the sputa, and the RNA was isolated. The RNA of sputum strains grown in culture was obtained in the same manner. An S. aureus-specific rRNA slot blot was developed to determine total RNA. Constant amounts of sample RNA were then spiked with serial dilutions of gene-specific competitor RNA for gyr, spa, hla, and RNAIII. After competitive RT-PCR the samples were separated by agarose gel electrophoresis and visualized after ethidium bromide staining. Quantification of transcripts was possible by comparison with the competitor. (A) Serial dilutions (1:1, 1:5, and 1:25) of samples were hybridized with a digoxigenin-labeled oligonucleotide specific for S. aureus 16S rRNA. Aliquots of RNA isolated in the exponential (exp) and the postexponential (p exp) growth phases of S. aureus isolates 21, 22, and 25 and aliquots of RNA isolated from sputum samples 21, 22, and 25 are shown. rDNA, rDNA PCR fragment starting with 10 fmol for the control dilution; c, control dilution (1:1). (B) Example of a gel after competitive RT-PCR. The top shows the detection of gyrase (gyr), and the bottom shows the detection of RNAIII. RNA was derived from the exponential (exp) and postexponential (p exp) growth phases of an S. aureus sputum strain and directly from the sputum. Constant amounts (1 and 0.1 ng) of sample RNA were spiked with serial dilutions of the competitors (100 to 0.16 amol and 1,000 to 1.6 amol, respectively; −, no competitor). In all three samples the same amount of gyrase was detectable. comp., competitor.