Fig. 5. 3D FISH reveals TCF-1 dependent TAD intermingling at the CD8a locus in DPs.
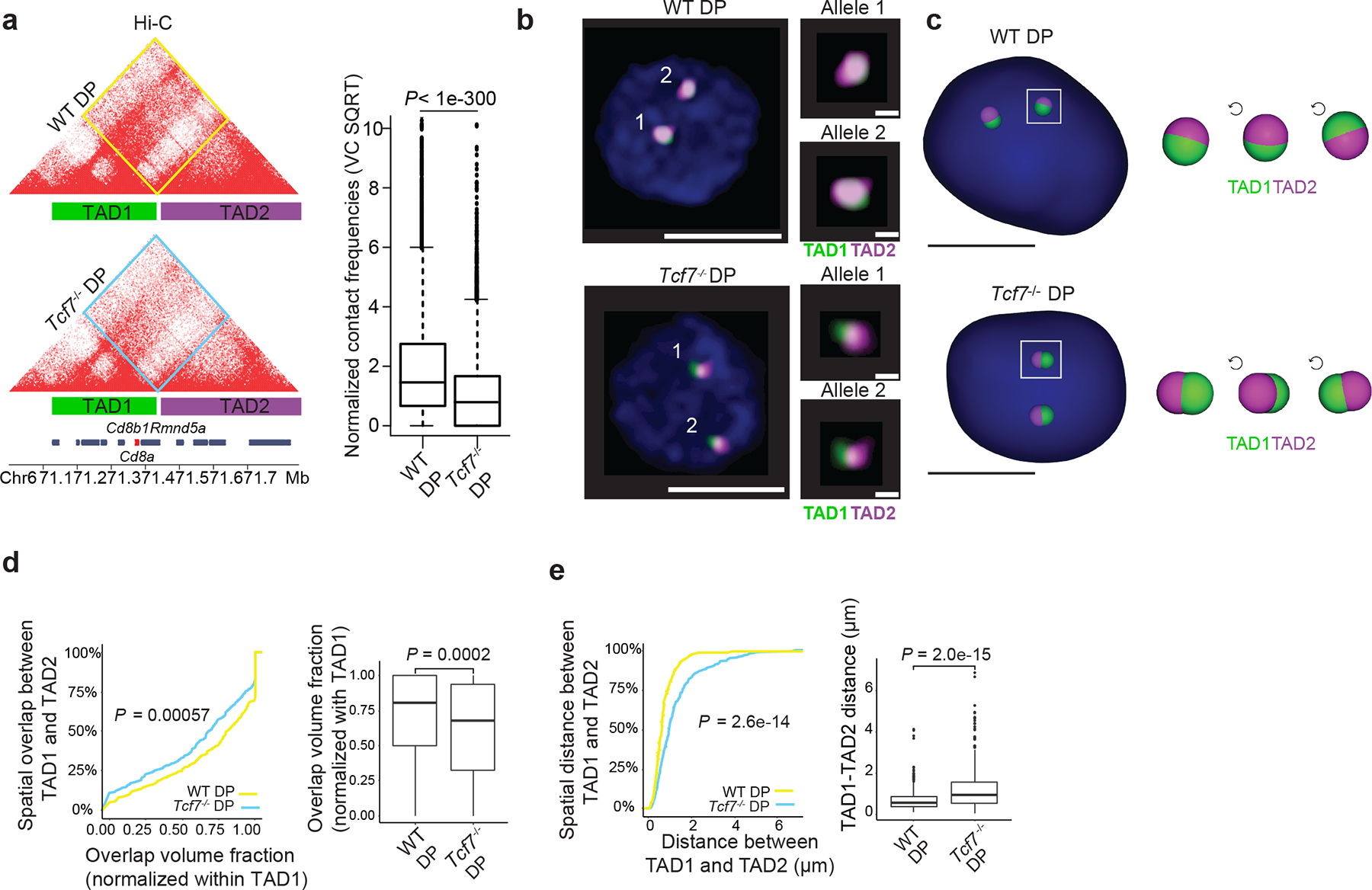
a. Contact matrix of Hi-C data at the Cd8a-Cd8b1 locus which includes the location of Oligopaint probes for TAD1 and TAD2 depicted as green and magenta bars. Inter-domain interactions are quantified by boxplot in wildtype and Tcf7−/− DPs (n=7221 genomic interactions; boxplot centre, median; box limits, upper (75th) and lower (25th) percentiles; whiskers, 1.5× interquartile range; points, outliers). Two sided Paired wilcoxon ranksum test was performed. TAD1 spans chr6:71163477-71432963 while TAD2 spans chr6:71440321-71825773 in mm10.
b. A representative image of Oligopaint FISH probes in wildtype and Tcf7−/− DPs with magnification of each allele. Scale bar for full cell is 5μm and for magnified allele 1 μm, nuclei are stained with DAPI (blue).
c. 3D rendering of TAD1 and TAD2 in wildtype and TCF-1 deficient single DP T cells. Zoomed view of one allele per cell is shown in 3 rotations of 90°, scale bar is 5μm.
d-e. Cumulative distribution plot (left) and corresponding boxplot (right) of overlap volume (d) and distance (e) between TAD1 and TAD2 across 312 individual wildtype DPs and 367 individual Tcf7−/− DPs. Kolmogorov-Smirnov test P-values and two-sided Wilcoxon rank-sum test P-values are shown for cumulative distribution plots and corresponding boxplots, respectively. Cells were pooled from two to three mice per genotype. Overlap volume was defined using a 3D segmentation strategy48 across a minimum of 300 alleles per condition. The overlap volume of TAD1 and TAD2 per allele was normalized to the volume of TAD1 (d). Distance was measured between the centroids of each domain across individual alleles (e). Data are shown as boxplots (centre, median; box limits, upper (75th) and lower (25th) percentiles; whiskers, 1.5× interquartile range; points, outliers)
