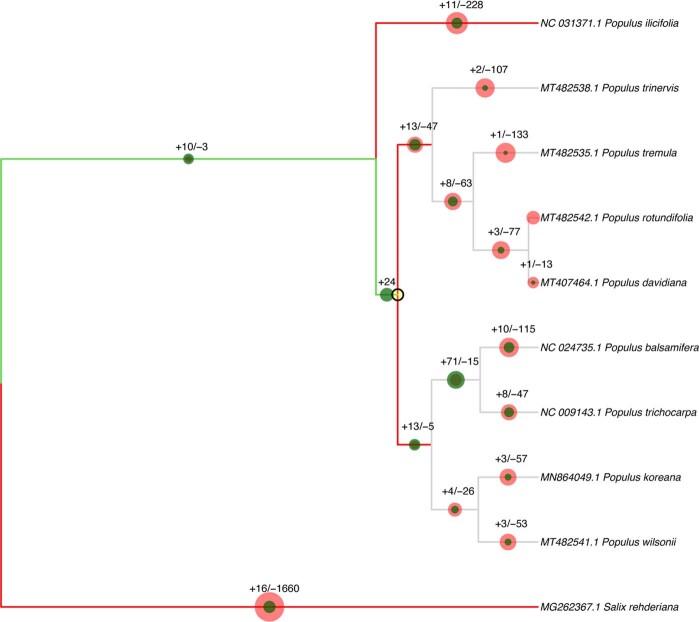
Extended Data Fig. 7. Phylogenetic placement results for our Populus chloroplast reads, using both transition and transversion SNPs, and using reads merged from all layers and sites.
The numbers on each edge represent the number of supporting (+) and conflicting (−) SNPs in the ancient Populus environmental genome overlapping the reference SNPs assigned to that edge. The ancient Populus environmental genome clearly contains a mixture of different species. The most likely placement is on the edge above Populus trichocarpa (NC 009143.1) and Populus balsamifera (NC 024735.1), with +71/−15 supporting and conflicting SNPs. However, we find some support for both branches directly leading to these species as well. Populus balsamifera and P. trichocarpa are considered sister species. They are both distributed in North America, as far North as Alaska, are known to hybridise both among themselves and other Populus species and are morphologically very similar93,99,100. Previous analyses found a very recent nuclear genome divergence time of only 75000 years ago for Populus trichocarpa and P. balsamifera100, but a deep chloroplast genome divergence time of at least 6-7 Ma99, which is an uncommon pattern in plants. Our ancient Populus sample could contain individuals either ancestral to, or hybridized from, the modern Populus trichocarpa and P. balsamifera species.