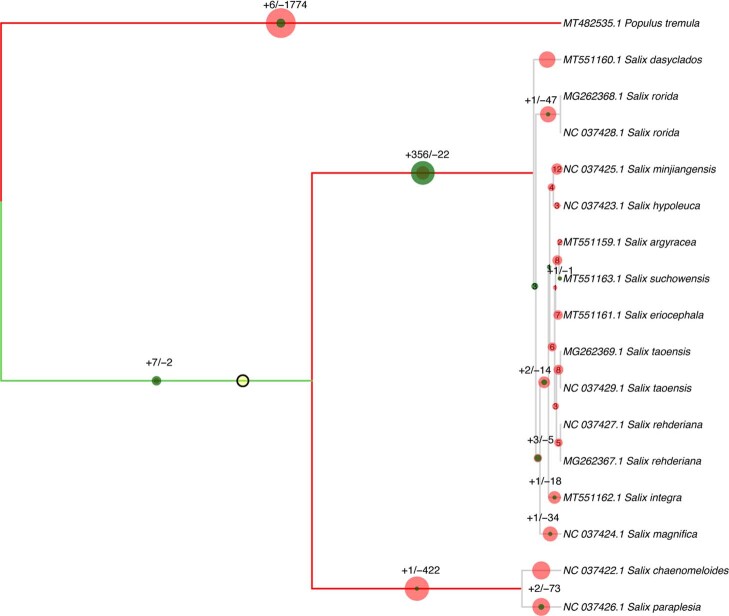
Extended Data Fig. 8. Phylogenetic placement results for our Salix chloroplast reads, using both transition and transversion SNPs, and using reads merged from all layers and sites.
The numbers on each edge represent the number of supporting (+) and conflicting (−) SNPs in the ancient Salix environmental genome overlapping the reference SNPs assigned to that edge. The ancient Salix environmental genome falls basal to a main Salix clade. Our ancient Salix sample is phylogenetically placed, with 356 supporting SNPs and 22 conflicting SNPs, on a basal branch leading to the main clade. Although the Salix chloroplast phylogeny is not considered fully resolved93, the difficulties in resolution lie underneath our placement branch, and this along with the high number of SNPs on the placement branch allow us to be confident in the placement position. Our chloroplast phylogeny agrees roughly with101, which estimated a divergence date between these two main Salix clades at 16.9 Ma, and a root age of the first clade, to which our ancient sample is basal to, of 8.1 Ma. It is reasonable, then, to conclude that our ancient Salix sample is at least 8.1 Ma diverged from modern Salix species, and probably represents an extinct species, or extant species without a reference genome sequenced, or a pool thereof.