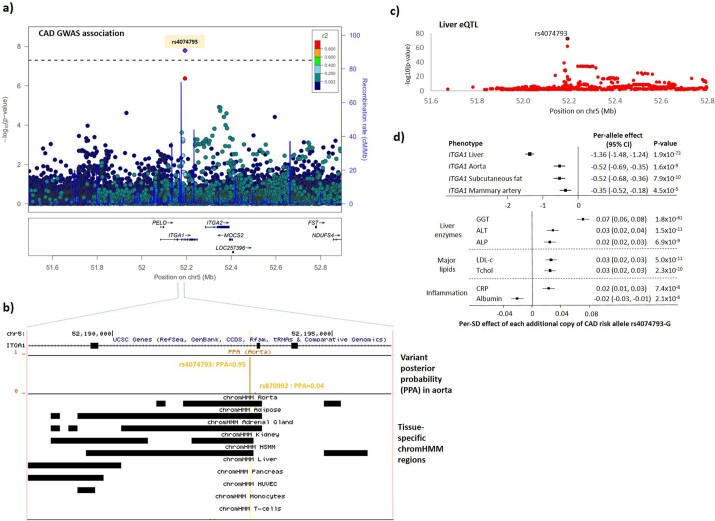
Extended Data Fig. 7. Prioritizing the likely causal variant, gene and pathway at the ITGA1 locus.
a, Regional association plot from the primary CAD meta-analysis for the ITGA1 region. Colored dots represent the position (x-axis) in GRCh37 coordinates and –log10(meta-analysis P value) (y-axis) of each variant in the region. Dots are shaded to represent the r2 with the lead CAD variant (rs4074793), estimated using a random sample of 5,000 European-ancestry participants from UK Biobank. Recombination peaks are plotted in blue based on estimates of recombination from 1000 Genomes European-ancestry individuals. b, Tissue-specific imputed chromHMM states at the two credible set variants in the ITGA1 region. The top track shows the position on chromosome 5 (GRCh37) with respect to the ITGA1 gene. The second track shows as a vertical orange line the posterior probability (y-axis) for each variant in the region from the FGWAS fine-mapping, identifying rs4074793 (PPA = 0.95) as the likely causal variant. The third track indicates as a black box the position of an enhancer state in each of the ten CAD-relevant tissues, using custom imputed chromHMM states based on epigenomic data from the NIH Roadmap Epigenomics Consortium project. The yellow vertical line indicates the position of the likely causal variant (rs4074793) with respect to the chromHMM states. rs4074793 is annotated to a chromHMM state for all five tissues that show enrichment in the region. HSMM, human skeletal muscle cells; HUVEC, human umbilical vein endothelial cells; PPA, posterior probability of being the causal variant. c, Effect of rs4074973 on ITGA1 expression in liver in the STARNET study. The plot shows the position (x-axis) in GRCh37 coordinates and –log10(P value) (y-axis) of each variant in the region. The likely causal CAD variant rs4074973 is circled in black. Only variants with P < 0.01 are displayed. d, Associations of rs4074973 with ITGA1 expression and phenotypes from a phenome-wide association study. The per-allele association of rs40747973-G (the CAD risk allele) measured in s.d. units is plotted for each phenotype. The box indicates the point estimate and the horizontal bars represent the 95% confidence intervals. The top panel shows the association estimates for ITGA1 expression from the STARNET study. The bottom panel shows associations from UK Biobank (liver enzymes and inflammatory markers) and the literature (lipids46). ALP, alkaline phosphatase; ALT, alanine aminotransferase; CRP, C-reactive protein; GGT, gamma glutamyltransferase; LDL-c, low-density lipoprotein cholesterol; Tchol, total cholesterol.