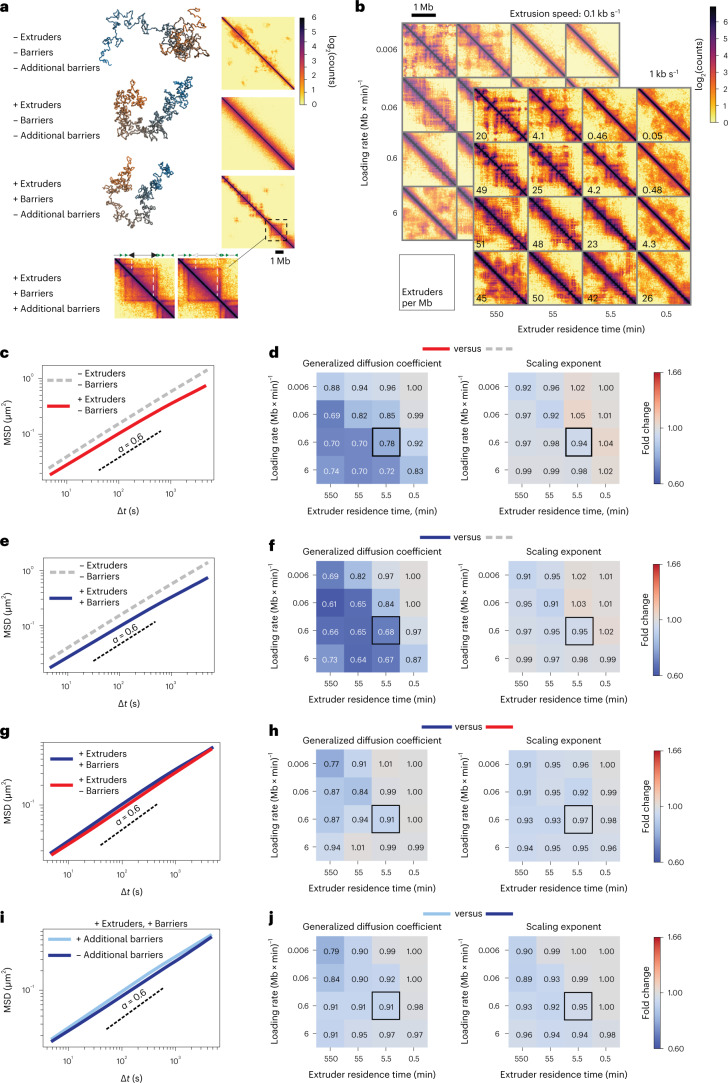
Fig. 2. Loop extrusion generally slows down polymer motion.
a, Representative snapshots of conformations and simulated contact maps for a polymer model with excluded volume and increasingly complex models with loop extruders, extrusion barriers sampled from CTCF motifs within 9 Mb on chromosome 15 (Chr15:7–16 Mb) and additional randomly distributed extrusion barriers. For the system with additional barriers, the contact map is presented aside with magnification of the contact map of the system without additional barriers to highlight the differences. b, Simulated contact maps (with loop extrusion and extrusion barriers) for polymers with two extrusion speeds (1 kb s−1 and 0.1 kb s−1) and different combinations of extruder loading rates and residence times. The resulting linear densities of extruders (number per Mb) are shown in the bottom left corner of each contact map. c, Effect of extruders. MSDs of polymers with (red line) or without (gray dashed line) loop extruders in the absence of extrusion barriers (loading rate 0.6 (Mb × min)−1 and residence time 5.5 min, corresponds to black square in panel d). Black dashed curve represents α = 0.6 as an eye guide. d, Effect of extruders. Ratios of generalized diffusion coefficients and anomalous exponents between the two conditions shown in panel c. Black square, set of parameters whose corresponding MSDs are shown in panel c. e, MSDs of polymers with (blue line) or without (gray dashed line) both extruders and barriers. Same parameters as in panel c. f, Same as panel d for cases illustrated in panel e. g, MSDs of polymers with loop extruders in the presence (blue) or absence (red) of extrusion barriers. Same parameters as in panels c and e. h, Same as panels d and f but for cases illustrated in panel g. i, MSDs of polymers either with (light blue) or without (red) additional randomly inserted extrusion barriers. Same parameters as in panels c, e, g. j, Same as panels d, f and h but for cases illustrated in panel i.