Fig. 1.
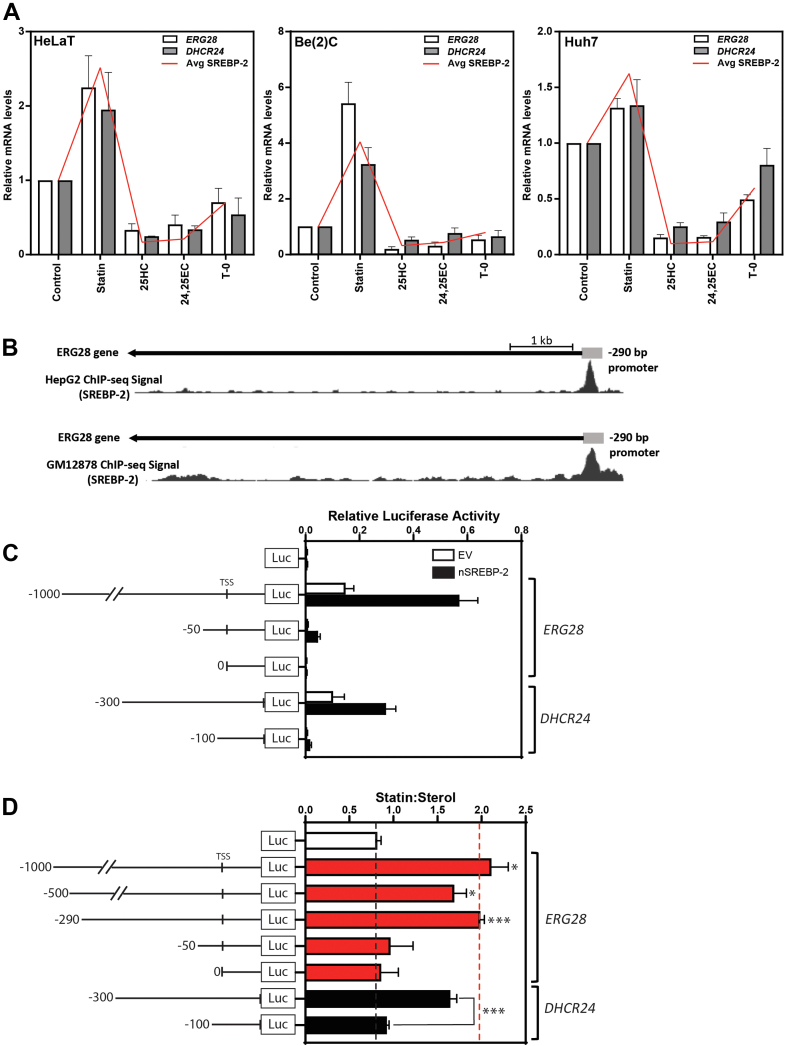
ERG28 is transcriptionally responsive to changes in sterol levels and responds like an SREBP-2 target. A: Previously generated cDNA sets (18, 19) from sterol-depleted Huh7, Be(2)C, and HeLaT cells treated for 24 h with 5 μM compactin (statin), 10 μM 25-hydroxycholesterol (25HC), 10 μM 24(S),25-epoxycholesterol (24,25EC), or 10 μM T0-901317 (T-0). mRNA levels for ERG28 and DHCR24 were measured using qRT-PCR and normalized to porphobilinogen deaminase (PBGD). mRNA levels are relative to the solvent control condition, which was set to 1. Data are presented as mean + SEM from n = 3 independent experiments, each performed in triplicate. The average responsiveness for SREBP-2 target genes already published from these cDNA sets (SQLE, LSS, and LDM (19) and HMGCR and TM7SF2 (18)) is represented by the red line (avg SREBP-2). B: ChIP-Seq signals from the UCSC Genome browser (20) of statin-treated HepG2 cells and rabbit IgG anti-SREBP-2 binding in the GM12878 lymphoblast cell line shows SREBP-2 binding in the ERG28 promoter with the −290/+10 bp region outlined. C and D: HeLaT cells were transfected for 24 h with 25 ng Renilla luciferase construct and 250 ng Firefly luciferase constructs as shown. C: Cells were in addition cotransfected with 25 ng nSREBP-2 or empty vector plasmid. Relative luciferase activity represents the ratio of luciferase:renilla activity. D: Cells were treated with 5 μM compactin (statin) or 10 μM 25HC (sterol) in sterol-depleted medium for 24 h and then harvested, and a luciferase assay was performed. The ratio of normalized statin to sterol luciferase activities is presented for each construct. Data are presented as mean + SEM from n = 3–6 independent experiments, each performed in triplicate cultures. ERG28 constructs were compared with luciferase:renilla values for the -luc only construct, and DHCR24 construct activity was compared between the -300/0 and -100/0 DHCR24-luc constructs. Statistical significance was determined using a paired two-tailed t-test (∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.005).