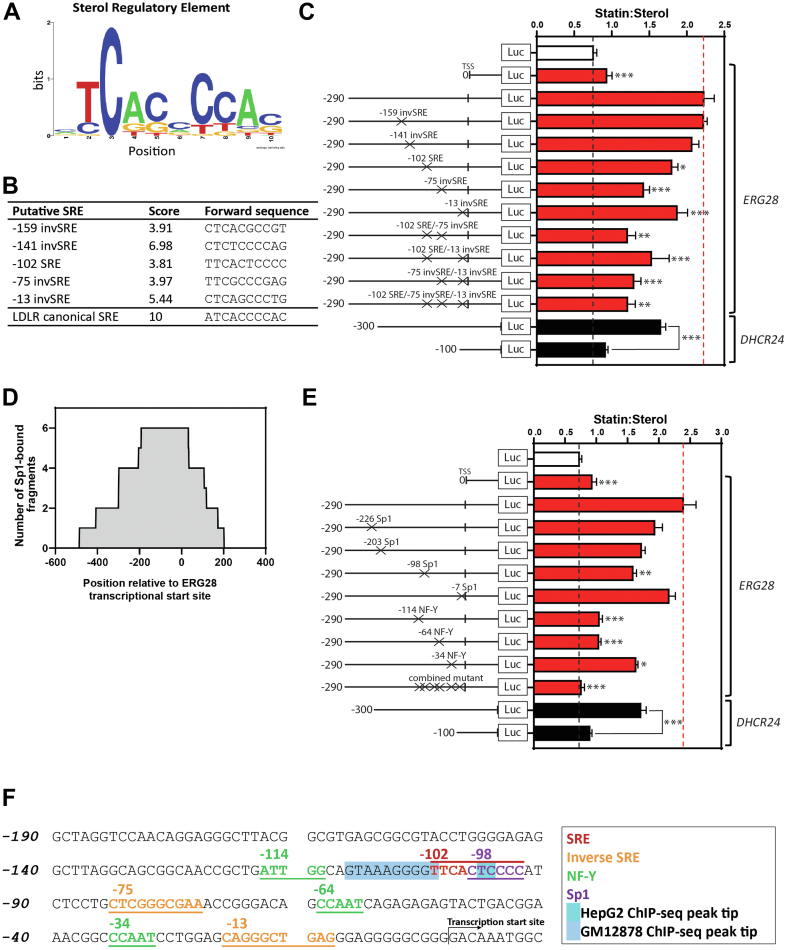
Fig. 2.
Mapping of SREs and other transcription factor binding sites in the ERG28 promoter. A: A weighted positional logo of the SREBP-2 SRE consensus sequence created using WebLogo (40) generated from our in-house matrix (41). B: The top five predicted SREs in the ERG28 promoter from our prediction FIMO (26). The canonical SRE found in the promoter of LDLR is included for reference and given the top available score of 10. C and E: Dual luciferase assay data identifying (C) SREs and (E) Sp1 and NF-Y binding sites in the ERG28 −290/+80 promoter. As for Fig. 1D, HeLaT cells were transfected for 24 h with 25 ng Renilla luciferase construct and 250 ng Firefly luciferase constructs as shown. Cells were treated with 5 μM compactin (statin) or 10 μM 25-HC (sterol) in sterol-depleted medium for 24 h and then harvested, and a luciferase assay was performed. The ratio of normalized statin to sterol luciferase activities is presented for each construct. Data are presented as mean + SEM from n = 4–8 separate experiments, each performed in triplicate cultures. Statistical significance was determined using a paired two-tailed t–test, and comparisons were between the wildtype −290/+80 ERG28-luc constructs and the respective SRE, Sp1, and NF-Y mutants and shown for significant decreases (∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.005). D: Combined ChIP-Seq signal graph showing binding of six Sp1-occupied sequence fragments taken from ENCODE project data (42) within the ERG28 −500 to +200 bp region. The Y-axis shows the number of times that each nucleotide was represented in a sequence fragment. F: A proposed map of transcriptional elements in the −190/+10 ERG28 promoter (compiled from data in Fig. 2C, E). The tips of the SREBP-2 ChIP-Seq peaks obtained from UCSC genome browser are highlighted in shades of blue.