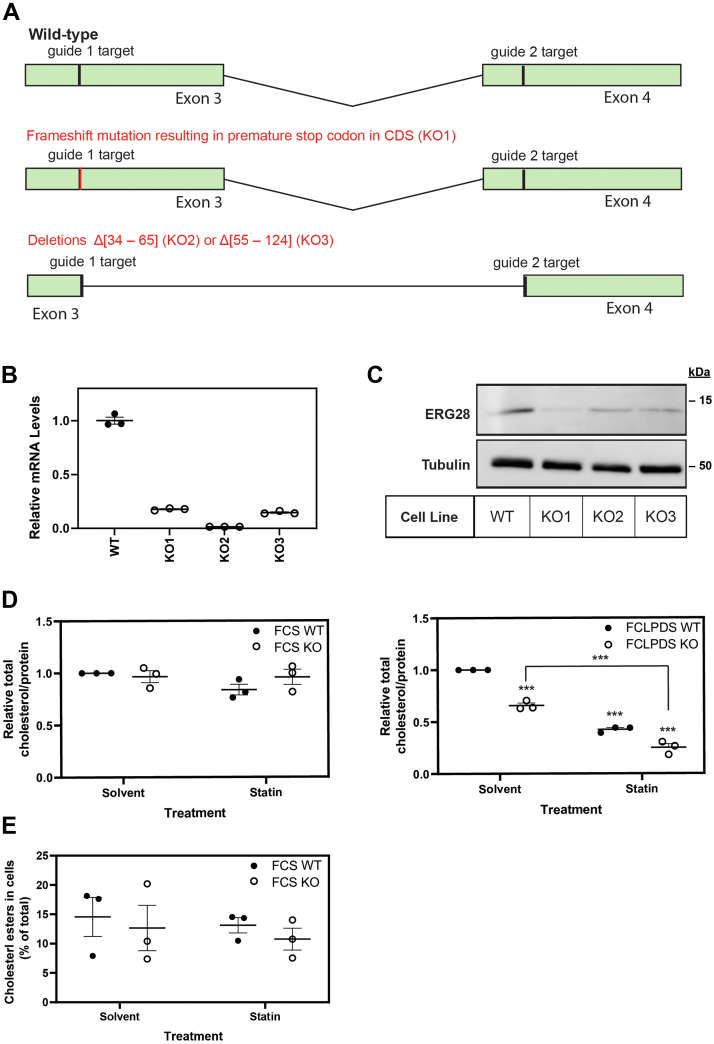
Fig. 4.
ERG28 KO cell lines have lower total cholesterol levels. A: CRISPR-Cas9 with two guides targeting exons 3 and 4 of human ERG28 was used to generate ERG28 KO cells in Huh7 cells. B: Total RNA was isolated from wild-type and KO cells, and ERG28 mRNA levels were measured using qRT-PCR. An overall reduction in ERG28 mRNA was observed in ERG28 KO cells. C: Protein lysates from ERG28 KO cells were normalized and separated by 12% SDS-PAGE for Western blotting. Membranes were probed for α-tubulin (for housekeeping) and endogenous ERG28 (nbp2-84865). D: Total cholesterol levels were measured in Huh7 wild-type and ERG28 KO cells using the Amplex Red Cholesterol Assay Kit (Invitrogen). Cells were plated in 10% FCLPDS DMEM:HG, and after 24 h, media were replaced and cells were grown for 48 h in either 10% FCS DMEM:HG or 10% FCLPDS DMEM:HG supplemented with 5 μM compactin (statin) or a DMSO solvent control. Cell lysates were harvested, and protein and total cholesterol levels were measured. Cholesterol levels were normalized to the concentration of protein and expressed relative to the wild-type solvent control cholesterol levels in each experiment, which was set to 1. E: Free and total cholesterol levels were measured in Huh7 wild-type and ERG28 KO cells using the Amplex Red Cholesterol Assay Kit (Invitrogen) as per D. Cells were plated in 10% FCLPDS DMEM:HG, and after 24 h, media were replaced and cells were grown for 48 h in either 10% FCS DMEM:HG supplemented with 5 μM compactin (statin) or a DMSO solvent control. Cell lysates were harvested, and protein and free and total cholesterol levels were measured. Cholesterol levels were normalized to the concentration of protein, and the ratio of free to total cholesterol was used to calculate the percentage of cholesterol stored as cholesteryl esters within the cell. Data are presented as the mean ± SEM from n = 3 independent experiments. Statistical significance from the solvent-treated wild-type was determined using a paired two-tailed t-test (∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.005).