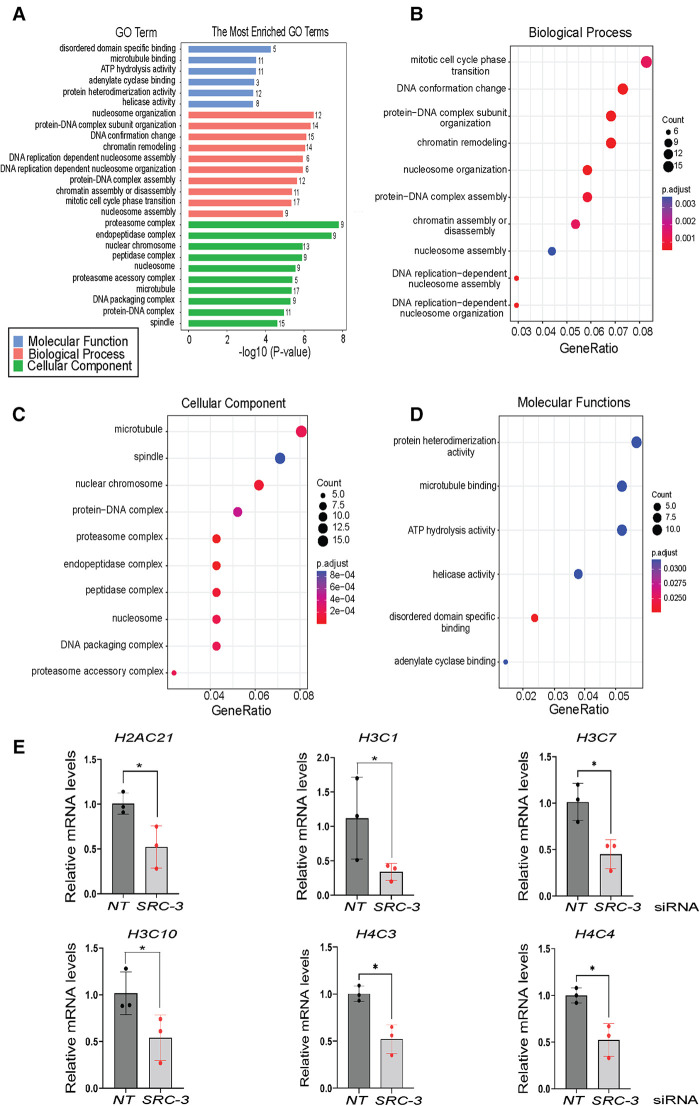
Figure 6.
Gene ontology functional analyses of differentially expressed genes in T-HESCs following SRC-3 knockdown (A) gene ontology (GO) enrichment analysis of differentially expressed genes was achieved using the DAVID. The six significantly enriched GO terms (p < 0.05) in molecular function along with the ten top significantly enriched GO terms in biological processes and cellular component branches are presented. The adjusted p-values of the terms were –log10 transformed. (B–D) Dot plots of enriched genes within the differentially expressed gene set are stratified according to biological processes, cellular components, and molecular functions respectively. (E) Quantitative real time PCR analysis shows significant reduction in the induction of the following histone family members: H2AC21, H3C1, H3C7, H3C10, H4C3, and H4C4 in T-HESCs following SRC-3 knockdown.