Fig. 2.
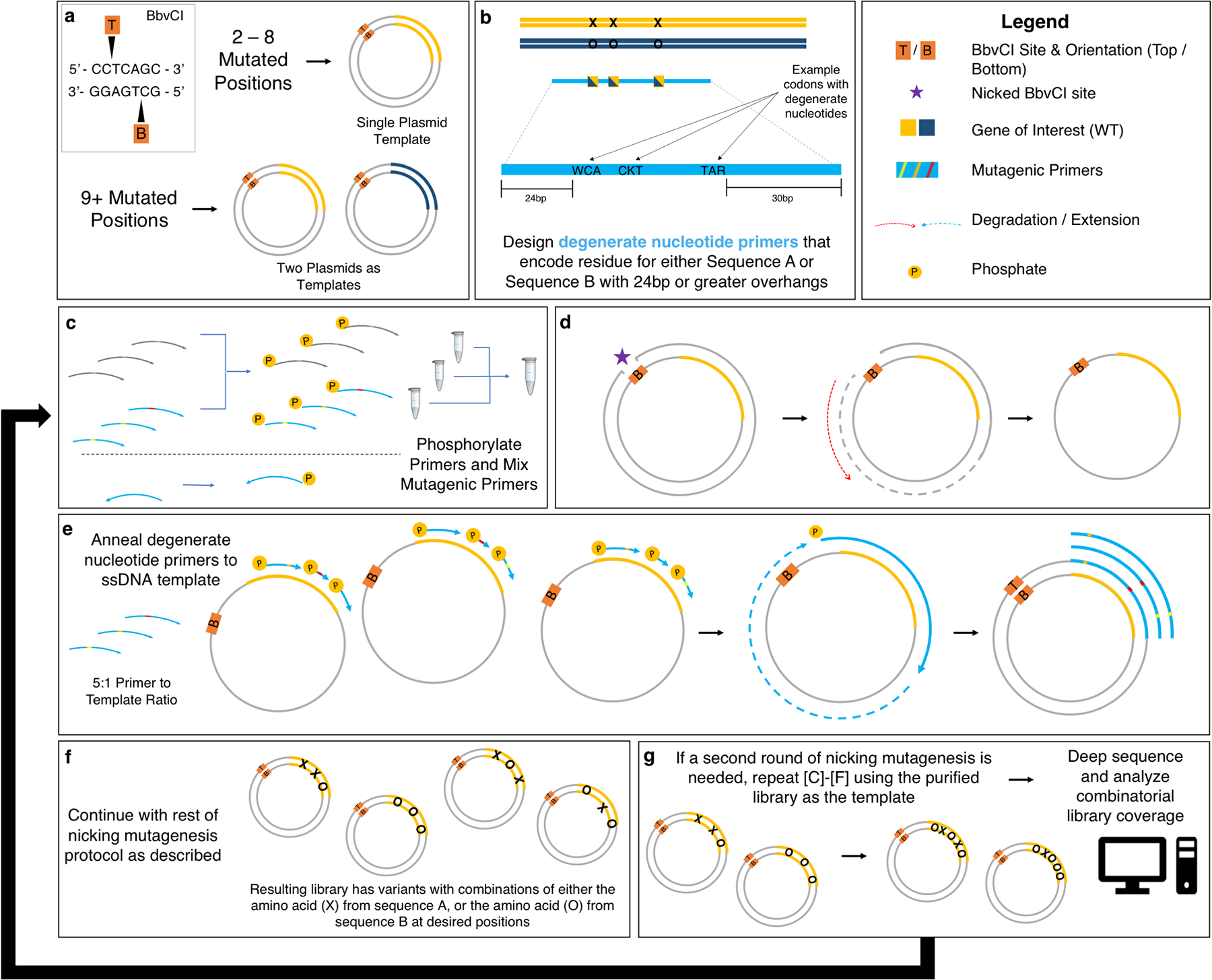
Flowchart for combinatorial mutagenesis. (a) Gene(s) of interest inserted into plasmid. One or two parental plasmids are needed depending on number of mutated positions desired. (b) Design of degenerate nucleotide primers to encode mutations in library. (c) Phosphorylate primers individually and mix together prior to mutagenesis. (d) Selectively nick and degrade one strand of plasmid DNA leaving circular ssDNA template. (e) Anneal mutagenic primers to template, extend around template and ligate to form heteroduplex DNA. (f) The remaining steps follow nicking mutagenesis protocol as previously described. (g) If a second round of nicking mutagenesis is required, repeat steps (c)-(f) using the purified library from the first round as the parental plasmid for the second round. The library can then be prepared for sequencing, sequenced, and analyzed.
