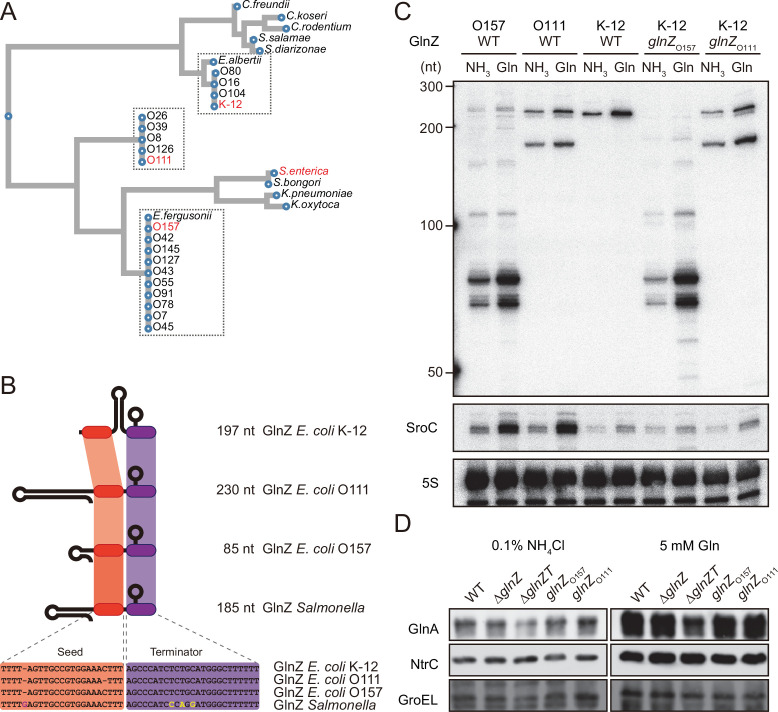
Figure 2. Variation of GlnZ sRNAs in the Enterobacteriaceae family.
(A) Multiple sequence alignment of glnA 3′UTRs using CLUSTALW program (https://www.genome.jp/tools-bin/clustalw). (B) The location of conserved seed region and Rho-independent terminator in different types of GlnZ sRNAs. The extra G nucleotide and variable nucleotides found in the terminator of Salmonella GlnZ are indicated in purple and yellow letters. (C) Escherichia coli strains express different types of GlnZ sRNAs. E. coli O157, O111, and K-12 strains were grown to exponential phase (OD600 ~0.5) in MOPS media containing 0.2% glucose as the carbon source and either 0.1% ammonium or 5 mM glutamine (Gln) as the nitrogen source. GlnZ and SroC sRNAs were detected by MMO-0416 and JVO-5622, respectively. 5S rRNA detected by MMO-1056 served as a loading control. (D) The difference in 3′UTR sequence does not affect the expression of downstream NtrC. E. coli K-12 strains, wild type (WT), ΔglnZ, ΔglnZT, glnZO157, and glnZO111, were grown to exponential phase (OD600 ~0.5) in MOPS media containing 0.2% glucose as the carbon source and either 0.1% ammonium or 5 mM Gln as the nitrogen source. GlnA was detected by an antibody raised against a synthetic peptide. NtrC was chromosomally tagged with 3xFLAG and detected by α-FLAG antibody. GroEL served as a loading control.