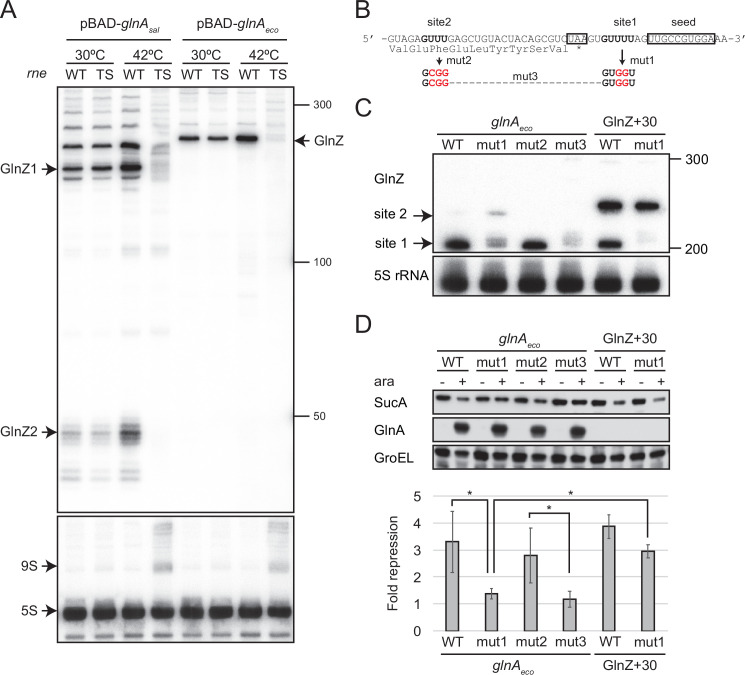
Figure 6. GlnZ release from the glnA mRNA is necessary for target repression.
(A) RNase E is essential for the processing of glnA mRNA. Escherichia coli ΔglnZ (wild-type [WT]) and ΔglnZ ams-1 (TS) strains harboring pBAD-glnAsal or pBAD-glnAeco were grown to OD600 ~0.5 at 30°C and split into two flasks. The flasks were incubated at either 30°C or 42°C for 30 min and further incubated for 10 min after adding 0.2% l-arabinose. The size is estimated by DynaMarker RNA Low II ssRNA fragment. (B) The nucleotide sequence of RNase E cleavage sites in E. coli K-12 glnA mRNA. The C-terminal amino acid sequence of GlnA is shown below the nucleotide sequence. The glnA stop codon and the GlnZ seed region are boxed. The mutated nucleotides are indicated in red. (C) The processing of GlnZ is abrogated by the mutations in RNase E cleavage sites. GlnZ processed from either the glnA mRNA or the GlnZ precursor sRNA was analyzed by northern blot. 5S rRNA served as a loading control. (D) The processing of GlnZ is required for the repression of SucA in the form of mRNA but not the precursor sRNA. Expression levels of SucA and GlnA were analyzed by western blot. GroEL served as a loading control. E. coli ΔglnZ strains harboring pBAD expression plasmids were grown to exponential phase (OD600 ~1.0) in LB medium in the absence (-) or presence of 0.01% l-arabinose (+). Bar graph represents the fold change of SucA repression by the glnA mRNA induced by arabinose calculated from biological replicates (n>5) with standard deviation. Statistical significance was calculated using one-way ANOVA and denoted as *p<0.05.