Figure 2.
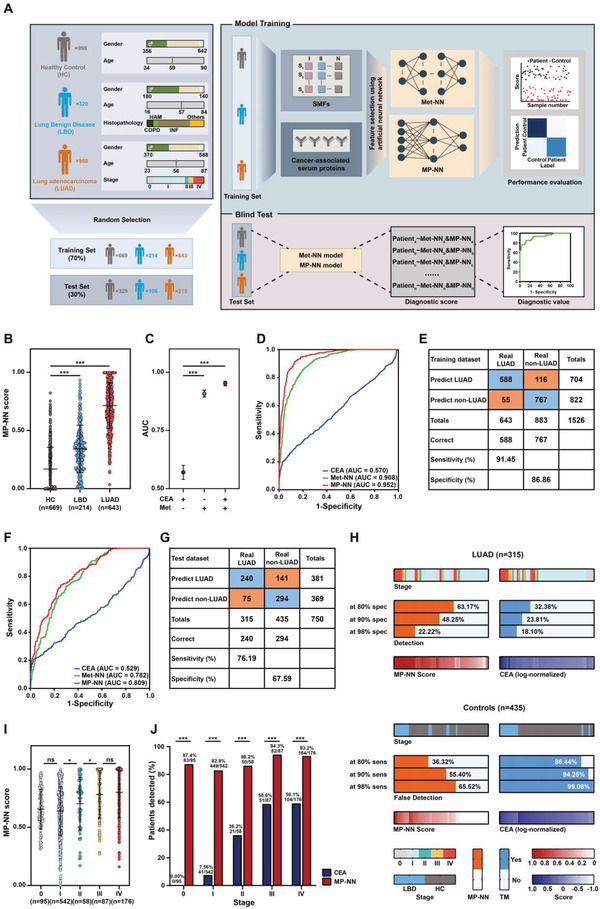
Development and blind test of serum metabolic fingerprints (SMFs) based lung adenocarcinoma (LUAD) diagnostic model. A) Schematic overview of the deep learning approach used to develop and validate the SMFs based integrated LUAD diagnostic model. B) Score of the MP‐NN identified in the training cohort. p‐values were calculated using a Wilcoxon test. Error bars refer to interquartile. C) Area‐under‐curve (AUC) for individual parameters in the training cohort. p‐values were calculated using a DeLong test. Error bars refer to 95% confidence intervals (CIs). D) Receiver operating characteristic curve (ROC) of the individual parameters in the training cohort. E) Confusion tables of binary results of the MP‐NN model in the training cohort. F) ROC of the individual parameters in the test cohort. G) Confusion tables of binary results of the MP‐NN model in the test cohort. H) Detection rates of MP‐NN and carcinoembryonic antigen (CEA) at different specificity in the test cohort. I) MP‐NN score levels summarized by stage in the whole LUAD cohort. p‐values were calculated using a Chi‐square test. Error bars indicate interquartile. J) MP‐NN detection rates summarized by stage in the whole LUAD cohort. p‐values were calculated using a Chi‐square test. *p < 0.05, and *** p < 0.001.
